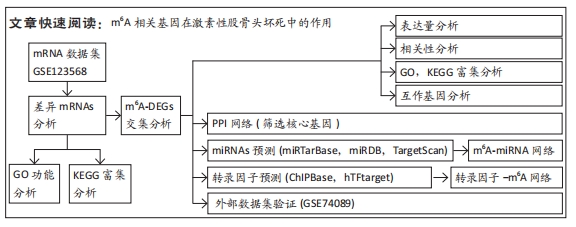
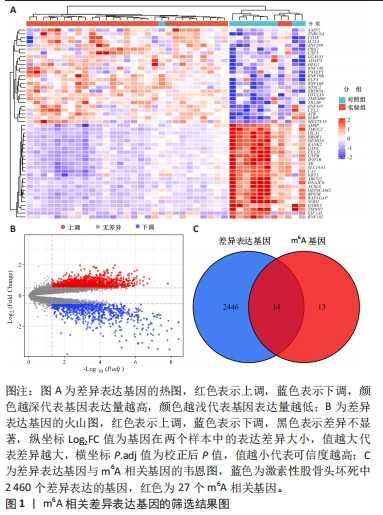
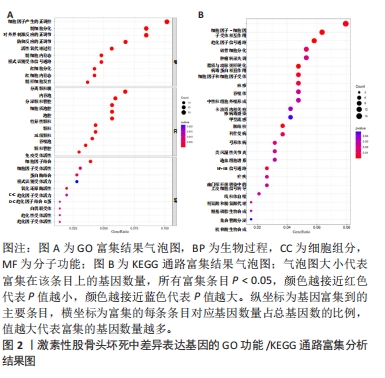
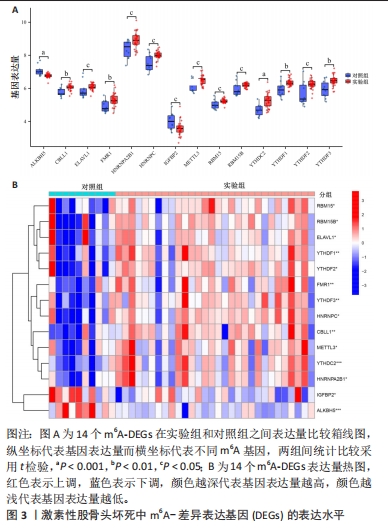
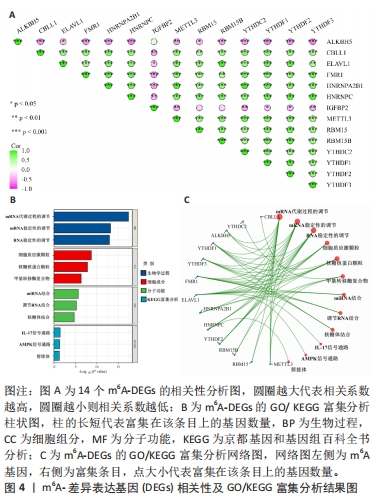
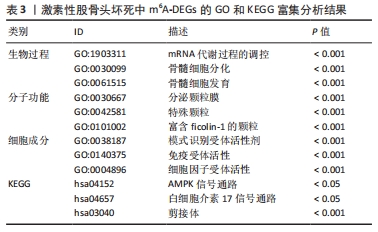
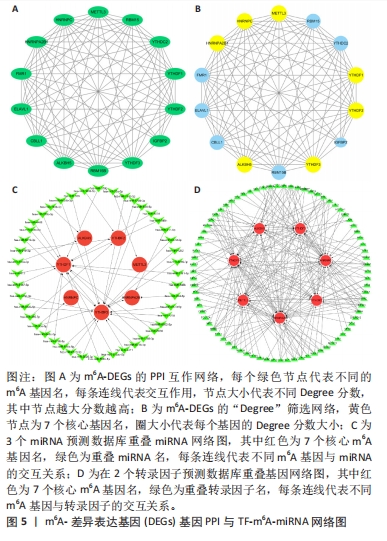
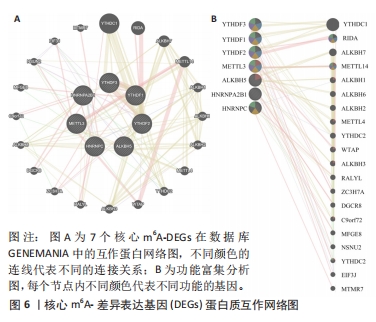
[1] FUKUSHIMA W, FUJIOKA M, KUBO T, et al. Nationwide epidemiologic survey of idiopathic osteonecrosis of the femoral head. Clin Orthop Relat Res. 2010;468(10): 2715-2724.
[2] GUERADO E, CASO E. The physiopathology of avascular necrosis of the femoral head: an update. Injury. 2016;47 Suppl 6:S16-S26.
[3] YUE J, YU H, LIU P, et al. Preliminary study of icariin indicating prevention of steroid-induced osteonecrosis of femoral head by regulating abnormal expression of miRNA-335 and protecting the functions of bone microvascular endothelial cells in rats. Gene. 2021;766:145128.
[4] HUANG C, WEN Z, NIU J, et al. Steroid-induced osteonecrosis of the femoral head: novel insight into the roles of bone endothelial cells in pathogenesis and treatment. Front Cell Dev Biol. 2021;9:777697.
[5] GUAN Q, LIN H, MIAO L, et al. Functions, mechanisms, and therapeutic implications of METTL14 in human cancer. J Hematol Oncol. 2022;15(1):13.
[6] 陈伟坚,张罡瑜,林天烨,等.RNA m6A甲基化修饰参与及调控骨科相关疾病[J].中国组织工程研究,2021,25(26):4236-4242.
[7] XU Y, YE S, ZHANG N, et al. The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun (Lond). 2020;40(10):484-500.
[8] CHENG C, ZHANG H, ZHENG J, et al. METTL14 benefits the mesenchymal stem cells in patients with steroid-associated osteonecrosis of the femoral head by regulating the m6A level of PTPN6. Aging (Albany NY). 2021;13(24):25903-25919.
[9] WANG Y, ZHANG Y, LU H, et al. Integrative analysis of cellular autophagy-related genes in steroid-induced osteonecrosis of the femoral head. Am J Transl Res. 2023; 15(9):5850-5872.
[10] CHAI JL, LU BW, DU HT, et al. Pyroptosis-related potential diagnostic biomarkers in steroid-induced osteonecrosis of the femoral head. BMC Musculoskelet Disord. 2023;24(1):609.
[11] LU H, FAN Y, YAN Q, et al. Identification and validation of ferroptosis-related biomarkers in steroid-induced osteonecrosis of the femoral head. Int Immunopharmacol. 2023;124(Pt A):110906.
[12] ZHOU R, PARK JW, CHUN RF, et al. Concerted effects of heterogeneous nuclear ribonucleoprotein C1/C2 to control vitamin D-directed gene transcription and RNA splicing in human bone cells. Nucleic Acids Res. 2017;45(2):606-618.
[13] WEN JR, TAN Z, LIN WM, et al. Role of m (6)A Reader YTHDC2 in differentiation of human bone marrow mesenchymal stem cells. Sichuan Da Xue Xue Bao Yi Xue Ban. 2021;52(3):402-408.
[14] BAI J, ZHANG W, ZHOU C, et al. MFG-E8 promotes osteogenic differentiation of human bone marrow mesenchymal stem cells through GSK3beta/beta-catenin signaling pathway. FASEB J. 2023;37(6):e22950.
[15] CHOI YJ, JEONG S, YOON KA, et al. Deficiency of DGCR8 increases bone formation through downregulation of miR-22 expression. Bone. 2017;103:287-294.
[16] 恽波,周灵杰,凡进,等.GIT1通过Notch信号通路促进骨髓细胞向破骨细胞分化[J]. 南京医科大学学报(自然科学版),2016,36(9):1036-1040.
[17] JIANG Y, ZHANG Y, CHEN W, et al. Correction to: achyranthes bidentata extract exerts osteoprotective effects on steroid-induced osteonecrosis of the femoral head in rats by regulating RANKL/RANK/OPG signaling. J Transl Med. 2021;19(1):208.
[18] LIU Y, SHAN H, ZONG Y, et al. IKKe in osteoclast inhibits the progression of methylprednisolone-induced osteonecrosis. Int J Biol Sci. 2021;17(5):1353-1360.
[19] WANG N, WANG L, YANG J, et al. Quercetin promotes osteogenic differentiation and antioxidant responses of mouse bone mesenchymal stem cells through activation of the AMPK/SIRT1 signaling pathway. Phytother Res. 2021. doi:10.1002/ptr.7010.LA.
[20] JIANG T, XIA C, CHEN X, et al. Melatonin promotes the BMP9-induced osteogenic differentiation of mesenchymal stem cells by activating the AMPK/beta-catenin signalling pathway. Stem Cell Res Ther. 2019;10(1):408.
[21] KIM HJ, SEO SJ, KIM JY, et al. IL-17 promotes osteoblast differentiation, bone regeneration, and remodeling in mice. Biochem Biophys Res Commun. 2020;524(4): 1044-1050.
[22] SUN J, DENG Y, SHI J, et al. MicroRNA‑542‑3p represses OTUB1 expression to inhibit migration and invasion of esophageal cancer cells. Mol Med Rep. 2020;21(1):35-42.
[23] ZHANG HM, KUANG S, XIONG X, et al. Transcription factor and microRNA co-regulatory loops: important regulatory motifs in biological processes and diseases. Brief Bioinform. 2015;16(1):45-58.
[24] TONG W, WENZE G, LIBING H, et al. Exploration of shared TF-miRNA‒mRNA and mRNA-RBP-pseudogene networks in type 2 diabetes mellitus and breast cancer. Front Immunol. 2022;13:915017.
[25] XIAO Z, ZHOU M, JI X, et al. Research progress of microRNA-21 in femoral head necrosis: a review. Asian J Surg. 2023;46(7):3003-3004.
[26] CUI Y, HUANG T, ZHANG Z, et al. The potential effect of BMSCs with miR-27a in improving steroid-induced osteonecrosis of the femoral head. Sci Rep. 2022;12(1):21051.
[27] ZHUANG Y, CHENG M, LI M, et al. Small extracellular vesicles derived from hypoxic mesenchymal stem cells promote vascularized bone regeneration through the miR-210-3p/EFNA3/PI3K pathway. Acta Biomater. 2022;150:413-426.
[28] CHEN Y, YU H, ZHU D, et al. miR-136-3p targets PTEN to regulate vascularization and bone formation and ameliorates alcohol-induced osteopenia. FASEB J. 2020; 34(4):5348-5362.
[29] NAN K, ZHANG Y, ZHANG X, et al. Exosomes from miRNA-378-modified adipose-derived stem cells prevent glucocorticoid-induced osteonecrosis of the femoral head by enhancing angiogenesis and osteogenesis via targeting miR-378 negatively regulated suppressor of fused (Sufu). Stem Cell Res Ther. 2021;12(1):331.
[30] LI G, LIU H, ZHANG X, et al. The protective effects of microRNA-26a in steroid-induced osteonecrosis of the femoral head by repressing EZH2. Cell Cycle. 2020; 19(5):551-566.
[31] CHEN Y, TANG B, JIANG W, et al. miR-486-5p attenuates steroid-induced adipogenesis and osteonecrosis of the femoral head via TBX2/P21 axis. Stem Cells. 2023;41(7):711-723.
[32] KONG L, ZUO R, WANG M, et al. Silencing microRNA-137-3p, which Targets RUNX2 and CXCL12 prevents steroid-induced osteonecrosis of the femoral head by facilitating osteogenesis and angiogenesis. Int J Biol Sci. 2020;16(4):655-670.
[33] LIMLAWAN P, MARGER L, DURUAL S, et al. Delivery of microRNA-302a-3p by APTES modified hydroxyapatite nanoparticles to promote osteogenic differentiation in vitro. BDJ Open. 2023;9(1):8.
[34] YUAN X, SHI L, GUO Y, et al. METTL3 regulates ossification of the posterior longitudinal ligament via the lncRNA XIST/miR-302a-3p/USP8 axis. Front Cell Dev Biol. 2021;9:629895.
[35] 杨礼庆,董成建,朱姝.成骨分化与修复调控转录因子Runx2在股骨头坏死组织中的表达:非随机平行对照临床观察试验方案[J].中国组织工程研究, 2016,20(17):24529-24534.
[36] LIU DD, ZHANG CY, LIU Y, et al. RUNX2 regulates osteoblast differentiation via the BMP4 signaling pathway. J Dent Res. 2022;101(10):1227-1237.
[37] WANG FS, CHEN YS, KO JY, et al. Bromodomain protein BRD4 accelerates glucocorticoid dysregulation of bone mass and marrow adiposis by modulating H3K9 and Foxp1. Cells. 2020;9(6):1500.
[38] TAI P, WUH, VAN WIJNEN AJ, et al. Genome-wide DNase hypersensitivity, and occupancy of RUNX2 and CTCF reveal a highly dynamic gene regulome during MC3T3 pre-osteoblast differentiation. PLoS One. 2017;12(11):e188056.
[39] YU S, GUO J, SUN Z, et al. BMP2-dependent gene regulatory network analysis reveals Klf4 as a novel transcription factor of osteoblast differentiation. Cell Death Dis. 2021;12(2):197.
|