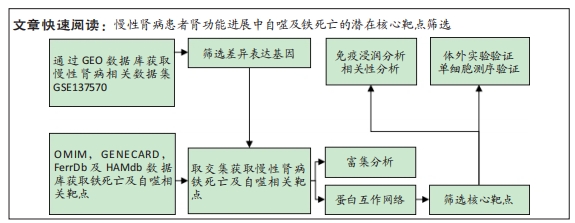
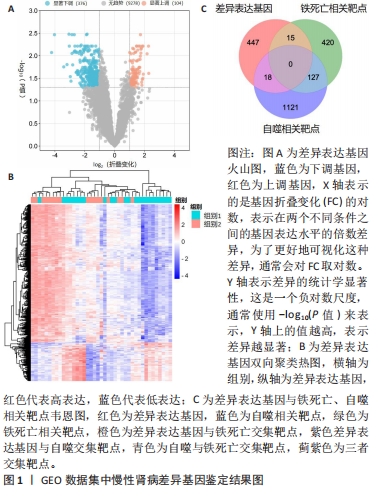
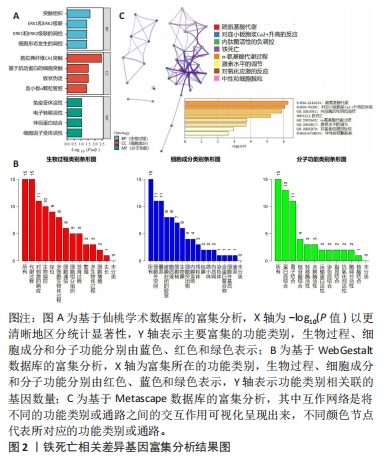
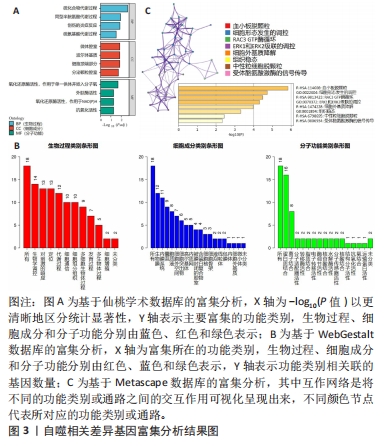
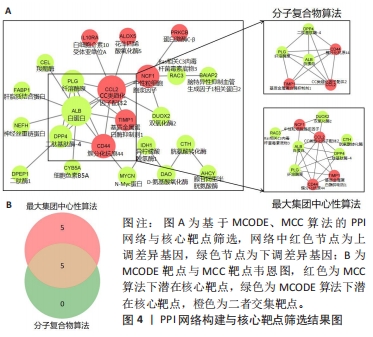
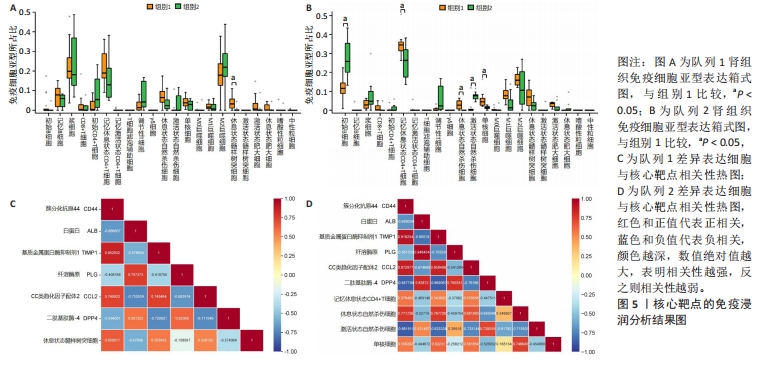
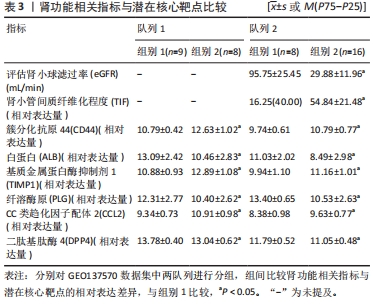
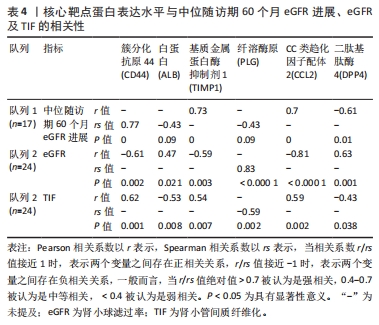
[1] 陈欠欠,朱凤阁,陈香美.姜黄素治疗肾间质纤维化分子机制研究进展[J].中草药,2023,54(3):966-975.
[2] HUMPHREYS BD. Mechanisms of renal fibrosis. Annu Rev Physiol. 2018;80(1): 309-326.
[3] JIANG X, STOCKWELL BR, CONRAD M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266-282.
[4] OTASEVIC V, VUCETIC M, GRIGOROV I, et al. Ferroptosis in different pathological contexts seen through the eyes of mitochondria. Oxid Med Cell Longev. 2021; 2021(6):5537330.
[5] WANG J, LIU Y, WANG Y, et al. The cross-link between ferroptosis and kidney diseases. Oxid Med Cell Longev. 2021;2021:6654887.
[6] 李磊,夏煜琦,程帆.铁死亡在肾纤维化中的作用研究进展[J].疑难病杂志, 2022,21(8):872-876.
[7] CAO W, LI J, YANG K, et al. An overview of autophagy: mechanism, regulation and research progress. Bull Cancer. 2021;108(3):304-322.
[8] CHOI EM. Autophagy in kidney disease. Annu Rev Physiol. 2020,82(1):297-322.
[9] LIU J, LIU Y, WANG Y, et al. TMEM164 is a new determinant of autophagy-dependent ferroptosis. Autophagy. 2023;19(3):945-956.
[10] CHUNG JJ, GOLDSTEIN L, Chen YJ, et al. Single-cell transcriptome profiling of the kidney glomerulus identifies key cell types and reactions to injury. J Am Soc Nephrol. 2020;31(10):2341-2354.
[11] PENG Z, GUO HY, LI YQ, et al. The Smad3-dependent microRNA let-7i-5p promoted renal fibrosis in mice with unilateral ureteral obstruction. Front Physiol. 2022;13:937878.
[12] KANG K, XIE F, MAO J, et al. Significance of tumor mutation burden in immune infiltration and prognosis in cutaneous melanoma. Front Oncol. 2020;10:573141.
[13] SORICE M. Crosstalk of autophagy and apoptosis. Cells. 2022;11(9):1479.
[14] 黄衍恒,叶霖,黄小荣,等.肾脏固有细胞自噬对肾纤维化作用的研究进展[J].中华肾脏病杂志,2022,38(3):247-253.
[15] PRERNA K, DUBEY VK. Beclin1-mediated interplay between autophagy and apoptosis: new understanding. Int J Biol Macromol. 2022;204:258-273.
[16] PENA-MARTINE C, RICKMAN AD, HECKMANN BL. Beyond autophagy: LC3-associated phagocytosis and endocytosis. Sci Adv. 2022;28;8(43):eabn1702.
[17] VARGAS JNS, HAMASAKI M, KAWABATA T, et al. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol. 2023;24(3):167-185.
[18] ZHANG B, CHEN X, RU F, et al. Liproxstatin-1 attenuates unilateral ureteral obstruction-induced renal fibrosis by inhibiting renal tubular epithelial cells ferroptosis. Cell Death Dis. 2021;12(9):843-843.
[19] GE MH, TIAN H, MAO L, et al. Zinc attenuates ferroptosis and promotes functional recovery in contusion spinal cord injury by activating Nrf2/GPX4 defense pathway. CNS Neurosci Ther. 2021;27(9):1023-1040.
[20] LI MY, DAI XH, YU XP, et al. Scalp acupuncture protects against neuronal ferroptosis by activating the p62-Keap1-Nrf2 pathway in rat models of intracranial haemorrhage. J Mol Neurosci. 2022;72(1):82-96.
[21] 米海潮,史敏,崔芳.铁自噬与铁死亡及其相关疾病[J].中国生物化学与分子生物学报,2022,38(9):1133-1140.
[22] JIN T, CHEN C. Umbelliferone delays the progression of diabetic nephropathy by inhibiting ferroptosis through activation of the Nrf-2/HO-1 pathway. Food Chem Toxicol. 2022;163:112892.
[23] WANG J, WANG Y, LIU Y, et al. Ferroptosis, a new target for treatment of renal injury and fibrosis in a 5/6 nephrectomy-induced CKD rat model. Cell Death Discov. 2022;8(1):127-127.
[24] ZHOU L, XUE X, HOU Q, et al. Targeting ferroptosis attenuates interstitial inflammation and kidney fibrosis. Kidney Dis (Basel). 2022;8(1):57-71.
[25] 张波,汝峰,陈湘,等.自噬通过抑制上皮间质转化减轻梗阻性肾病肾纤维化[J].中南大学学报(医学版),2021,46(6):601-608.
[26] 毛稳,夏琳颖,刘贤莉,等.三七皂苷通过抑制自噬减轻TGF-β1诱导的肾小管上皮细胞损伤[J].中国实验方剂学杂志,2022,28(20):86-91.
[27] RUBY M, GIFFORD CC, PANDEY R, et al. Autophagy as a therapeutic target for chronic kidney disease and the roles of TGF-β1 in autophagy and kidney fibrosis. Cells. 2023;12(3):412.
[28] DJUDJAJ S, BOOR P. Cellular and molecular mechanisms of kidney fibrosis. Mol Aspects Med. 2019;65:16-36.
[29] HOU W, XIE Y, SONG X, et.al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12(8):1425-1428.
[30] LI J, CAO F, YIN HL, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11(2):88.
[31] ZHANG L, YANG P, CHEN J, et al. CD44 connects autophagy decline and ageing in the vascular endothelium. Nat Commun. 2023;14(1):5524-5524.
[32] BAI RJ, Liu D, Li YS, et al. OPN inhibits autophagy through CD44, integrin and the MAPK pathway in osteoarthritic chondrocytes. Front Endocrinol (Lausanne). 2022;13:919366.
[33] 高翔,梅长林.《慢性肾病早期筛查、诊断及防治指南(2022年版)》解读[J].中国实用内科杂志,2022,42(9):735-739.
[34] WARD ES, GELINAS D, DREESEN E, et al. Clinical significance of serum albumin and implications of FcRn inhibitor treatment in IgG-mediated autoimmune disorders. Front Immunol. 2022;13:892534.
[35] HUANG H, LIN Q, Dai X, et al. Derivation and validation of urinary TIMP-1 for the prediction of acute kidney injury and mortality in critically ill children. J Transl Med. 2022;20(1):102.
[36] KOKENY G, NEMETH A, Kopp JB, et al. Susceptibility to kidney fibrosis in mice is associated with early growth response-2 protein and tissue inhibitor of metalloproteinase-1 expression. Kidney Int. 2022;102(2):337-354.
[37] WANG L, WANG J, CHEN L. TIMP1 represses sorafenib-triggered ferroptosis in colorectal cancer cells by activating the PI3K/Akt signaling pathway. Immunopharmacol Immunotoxicol. 2023;45(4):419-425.
[38] HEISSIG B, SALAMA Y, TAKAHASHI S, et al.The multifaceted role of plasminogen in inflammation. Cell Signal. 2020;75:109761.
[39] KERAGALA CB, MEDCALF RL. Plasminogen: an enigmatic zymogen. Blood. 2021; 137(21):2881-2889.
[40] TYKHOMYROV AA, NEDZVETSKY VS, AGCA CA, et al. Plasminogen/plasmin affects expression of glycolysis regulator TIGAR and induces autophagy in lung adenocarcinoma A549 cells. Exp Oncol. 2020;42(4):270-276.
[41] LIN Z, SHI JL, CHEN M, et al. CCL2: an important cytokine in normal and pathological pregnancies: a review. Front Immunol. 2022;13:1053457.
[42] 李秀萍,王玲,王馨,等.CC类趋化因子配体2对结核分枝杆菌感染人单核巨噬细胞THP-1自噬的影响及分子机制[J].临床误诊误治,2021,34(9):101-106.
[43] FANG S, TANG H, LI MZ, et al. Identification of the CCL2 PI3K/Akt axis involved in autophagy and apoptosis after spinal cord injury. Metab Brain Dis. 2023;38(4): 1335.
[44] 张志威,汪锴,姜长涛.菌源同工酶分析揭示菌源DPP4同工酶是潜在的抗2型糖尿病靶点[J].遗传,2023,45(8):629-631.
[45] CUI X, YUN X, SUN M, et al. HMGCL-induced β-hydroxybutyrate production attenuates hepatocellular carcinoma via DPP4-mediated ferroptosis susceptibility. Hepatol Int. 2023;17(2):377-392.
|