中国组织工程研究 ›› 2019, Vol. 23 ›› Issue (27): 4269-4274.doi: 10.3969/j.issn.2095-4344.1371
• 肌肉肌腱韧带组织构建 tissue construction of the muscle, tendon and ligament • 上一篇 下一篇
脊髓损伤后肌肉萎缩基因谱的生物信息学分析
黄 晖,王广积
- (海南省人民医院骨科,海南省海口市 570311)
Bioinformatics analysis of gene spectrum of muscle atrophy after spinal cord injury
Huang Hui, Wang Guangji
- (Department of Orthopedics, Hainan Provincial People’s Hospital, Haikou 570311, Hainan Province, China)
摘要:
文章快速阅读:
.jpg) 文题释义:
生物信息学:作为一门新的学科领域,它是把基因组DNA序列信息分析作为源头,在获得蛋白质编码区的信息后进行蛋白质空间结构模拟和预测,然后依据特定蛋白质的功能进行必要的药物设计。基因组信息学、蛋白质空间结构模拟以及药物设计构成了生物信息学的3个重要组成部分。从生物信息学研究的具体内容上看,生物信息学应包括这3个主要部分:①新算法和统计学方法研究;②各类数据的分析和解释;③研制有效利用和管理数据新工具。
脊髓损伤后肌肉萎缩:人们普遍认为由于脊髓损伤而瘫痪的肌肉会发生萎缩并出现肌肉力量的减弱。脊髓损伤后发生肌肉纤维体积的减小或数量的减少,可能是去神经支配萎缩或失用性萎缩。去神经支配萎缩是由于脊髓运动神经元或腹侧根部的运动神经损伤引起的;失用性萎缩是由于中枢和节段性突触驱动对存活的脊柱运动神经元的破坏导致肌肉激活丧失。
文题释义:
生物信息学:作为一门新的学科领域,它是把基因组DNA序列信息分析作为源头,在获得蛋白质编码区的信息后进行蛋白质空间结构模拟和预测,然后依据特定蛋白质的功能进行必要的药物设计。基因组信息学、蛋白质空间结构模拟以及药物设计构成了生物信息学的3个重要组成部分。从生物信息学研究的具体内容上看,生物信息学应包括这3个主要部分:①新算法和统计学方法研究;②各类数据的分析和解释;③研制有效利用和管理数据新工具。
脊髓损伤后肌肉萎缩:人们普遍认为由于脊髓损伤而瘫痪的肌肉会发生萎缩并出现肌肉力量的减弱。脊髓损伤后发生肌肉纤维体积的减小或数量的减少,可能是去神经支配萎缩或失用性萎缩。去神经支配萎缩是由于脊髓运动神经元或腹侧根部的运动神经损伤引起的;失用性萎缩是由于中枢和节段性突触驱动对存活的脊柱运动神经元的破坏导致肌肉激活丧失。
.jpg) 文题释义:
生物信息学:作为一门新的学科领域,它是把基因组DNA序列信息分析作为源头,在获得蛋白质编码区的信息后进行蛋白质空间结构模拟和预测,然后依据特定蛋白质的功能进行必要的药物设计。基因组信息学、蛋白质空间结构模拟以及药物设计构成了生物信息学的3个重要组成部分。从生物信息学研究的具体内容上看,生物信息学应包括这3个主要部分:①新算法和统计学方法研究;②各类数据的分析和解释;③研制有效利用和管理数据新工具。
脊髓损伤后肌肉萎缩:人们普遍认为由于脊髓损伤而瘫痪的肌肉会发生萎缩并出现肌肉力量的减弱。脊髓损伤后发生肌肉纤维体积的减小或数量的减少,可能是去神经支配萎缩或失用性萎缩。去神经支配萎缩是由于脊髓运动神经元或腹侧根部的运动神经损伤引起的;失用性萎缩是由于中枢和节段性突触驱动对存活的脊柱运动神经元的破坏导致肌肉激活丧失。
文题释义:
生物信息学:作为一门新的学科领域,它是把基因组DNA序列信息分析作为源头,在获得蛋白质编码区的信息后进行蛋白质空间结构模拟和预测,然后依据特定蛋白质的功能进行必要的药物设计。基因组信息学、蛋白质空间结构模拟以及药物设计构成了生物信息学的3个重要组成部分。从生物信息学研究的具体内容上看,生物信息学应包括这3个主要部分:①新算法和统计学方法研究;②各类数据的分析和解释;③研制有效利用和管理数据新工具。
脊髓损伤后肌肉萎缩:人们普遍认为由于脊髓损伤而瘫痪的肌肉会发生萎缩并出现肌肉力量的减弱。脊髓损伤后发生肌肉纤维体积的减小或数量的减少,可能是去神经支配萎缩或失用性萎缩。去神经支配萎缩是由于脊髓运动神经元或腹侧根部的运动神经损伤引起的;失用性萎缩是由于中枢和节段性突触驱动对存活的脊柱运动神经元的破坏导致肌肉激活丧失。摘要
背景:脊髓损伤后常伴随肌肉萎缩的发生,但是它的基本机制仍然不是十分清楚。
目的:旨在探讨脊髓损伤后肌肉萎缩的分子生物学机制。
方法:分析基因表达数据库中的脊髓损伤后肌肉萎缩的基因谱GSE45550。基因表达谱GSE45550包括对照组(脊髓损伤前)、实验组1(脊髓损伤后3 d)、实验组2(脊髓损伤后8 d)、实验组3(脊髓损伤后14 d)。组织为SD大鼠的比目鱼肌,每组6例。随后对4组样本数据进行差异基因分析、GO分析、通路分析。
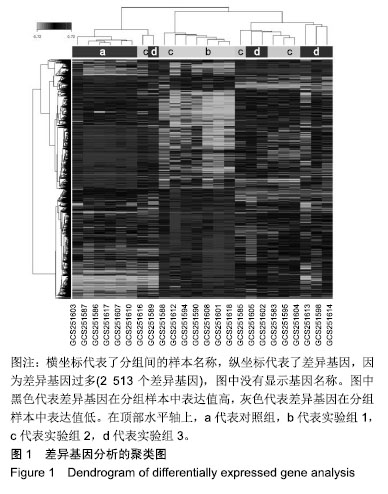
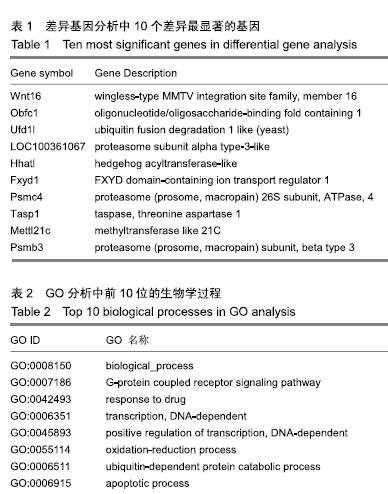
结果与结论: 确定了2 513个差异表达基因,其中Wnt16、Obfc1、Ufd1l、LOC100361067、Hhatl、Fxyd1、Psmc4、Tasp1、Mettl21c、Ufd1l差异表达最显著。GO分析显示差异基因的主要生物学过程为biological_process、G蛋白偶联受体信号通路、对药物的反应、DNA依赖性转录、DNA依赖性转录的正调节、氧化还原过程、泛素依赖性蛋白分解代谢过程、凋亡过程、RNA聚合酶转录的正调控及脂肪酸β-氧化。信号通路如MAPK信号、细胞凋亡、柠檬酸循环(TCA循环)可能起到重要的作用。研究比较完整地揭示了脊髓损伤后肌肉萎缩基因谱的差异表达基因和所涉及的生物学过程和信号通路,其中Wnt16可能是脊髓损伤后肌肉萎缩中的关键基因,为未来的治疗进展提供分子靶点。
中图分类号:
.jpg) 文题释义:
生物信息学:作为一门新的学科领域,它是把基因组DNA序列信息分析作为源头,在获得蛋白质编码区的信息后进行蛋白质空间结构模拟和预测,然后依据特定蛋白质的功能进行必要的药物设计。基因组信息学、蛋白质空间结构模拟以及药物设计构成了生物信息学的3个重要组成部分。从生物信息学研究的具体内容上看,生物信息学应包括这3个主要部分:①新算法和统计学方法研究;②各类数据的分析和解释;③研制有效利用和管理数据新工具。
脊髓损伤后肌肉萎缩:人们普遍认为由于脊髓损伤而瘫痪的肌肉会发生萎缩并出现肌肉力量的减弱。脊髓损伤后发生肌肉纤维体积的减小或数量的减少,可能是去神经支配萎缩或失用性萎缩。去神经支配萎缩是由于脊髓运动神经元或腹侧根部的运动神经损伤引起的;失用性萎缩是由于中枢和节段性突触驱动对存活的脊柱运动神经元的破坏导致肌肉激活丧失。
文题释义:
生物信息学:作为一门新的学科领域,它是把基因组DNA序列信息分析作为源头,在获得蛋白质编码区的信息后进行蛋白质空间结构模拟和预测,然后依据特定蛋白质的功能进行必要的药物设计。基因组信息学、蛋白质空间结构模拟以及药物设计构成了生物信息学的3个重要组成部分。从生物信息学研究的具体内容上看,生物信息学应包括这3个主要部分:①新算法和统计学方法研究;②各类数据的分析和解释;③研制有效利用和管理数据新工具。
脊髓损伤后肌肉萎缩:人们普遍认为由于脊髓损伤而瘫痪的肌肉会发生萎缩并出现肌肉力量的减弱。脊髓损伤后发生肌肉纤维体积的减小或数量的减少,可能是去神经支配萎缩或失用性萎缩。去神经支配萎缩是由于脊髓运动神经元或腹侧根部的运动神经损伤引起的;失用性萎缩是由于中枢和节段性突触驱动对存活的脊柱运动神经元的破坏导致肌肉激活丧失。