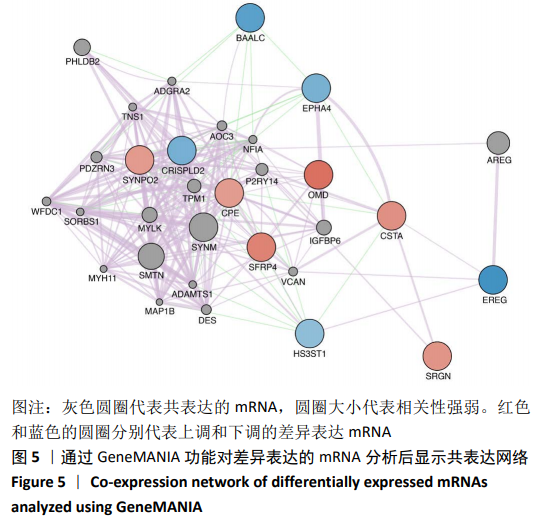
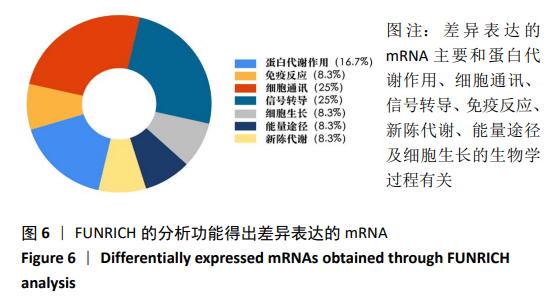
[1] 夏聪敏,许波,李刚,等.基于网络药理学探讨桂枝芍药知母汤治疗骨性关节炎的分子机制[J].中华中医药学刊,2018,36(11): 2681-2684.
[2] 李嘉程,许波,李刚,等.基于网络药理学研究杜仲抗骨质疏松的分子机制[J].中国现代中药,2018,20(8):28-34.
[3] 王利群,康坤丽.膝关节骨性关节炎中医治疗进展[J].中医临床研究,2015,15(15):114-117.
[4] 何仁豪.骨性关节炎发病机制及与microRNA相关性研究进展[J].中华实用诊断与治疗杂志,2012,26(11):1045-1047.
[5] FANG XN, YIN M, LI H, et al. Comprehensive analysis of competitive endogenous RNAs network associated with head and neck squamous cell carcinoma. Sci Rep. 2018;8(1):10544.
[6] JIANG H, MA R, ZOU S, et al. Reconstruction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in rheumatoid arthritis. Mol Biosyst. 2017;13(6):1182.
[7] SUI J, LI YH, ZHANG YQ, et al. Integrated analysis of long non-coding RNA associated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol. 2016;49(5):2023.
[8] CHOU CH, WU CC, SONG IW, et al. Genome-wide expression profiles of subchondral bone in osteoarthritis. Arthritis Res Ther. 2013;15(6):R190.
[9] SUN Y, MAUERHAN DR, HONEYCUTT PR, et al. Analysis of meniscal degeneration and meniscal gene expression. BMC Musculoskeletal Disorders. 2010;11(1):19-20.
[10] LI JH, LIU S , ZHOU H, et al. Starbase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale clip-seq data. Nucleic Acids Res. 2013;42(Database issue):D92-D97.
[11] CHOU CH, CHANG NW, SHRESTHA S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(Database issue):D239-D247.
[12] WONG N, WANG X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015; 43(Database issue):D146-D152.
[13] GENG C, ZIYUN W, DONGQING W, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013; 41(Database issue):D983-D986.
[14] MONTOJO J, ZUBERI K, RODRIGUEZ H, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. 2010;26(22): 2927.
[15] BARDOU P, MARIETTE J, ESCUDIÉ F, et al. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15(1):293.
[16] NAKANISHI R, AKIYAMA H, KIMURA H, et al. Osteoblast-Targeted Expression of Sfrp4 in Mice Results in Low Bone Mass. J Bone Miner Res. 2008;23(2):271-277.
[17] YANG L, DINARVAND P, QURESHI SH, et al. Engineering D-helix of antithrombin in alpha-1-proteinase inhibitor confers antiinflammatory properties on the chimeric serpin. Thromb Haemost. 2014;112(7): 164-175.
[18] 马剑锋,张艳彩,孙慧.下调SRGN基因表达对乳腺癌细胞凋亡及JAK/STAT信号通路的影响[J].癌症进展,2018,16(6):698-701.
[19] 许丁文,熊彦,严慧深,等.基于生物信息学分析丝甘蛋白聚糖对卵巢癌耐药性的影响及作用机制[J].中国药房,2019,30(1):46-51.
[20] SALMENA L, POLISENO L, TAY Y, et al. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell. 2011;146(3):358.
[21] BALLANTYNE MD, MCDONALD R, BAKER A. lncRNA/MicroRNA interactions in the vasculature. Clin Pharm Ther. 2016;99(5):494-501.
[22] 李西海,刘献祥.骨关节炎的核心病机—本痿标痹[J].中医杂志, 2014,55(14):1248-1249.
[23] 郑晓芬.骨关节炎发病机制和治疗的最新进展[J].中国组织工程研究,2017,21(20):3255-3262.
[24] SAXTON RA, SABATINI DM. mTOR Signaling in growth, metabolism, and disease. Cell. 2017;168(6):960-976.
[25] LAPLANTE M, SABATINI DM. mTOR signaling at a glance. J Cell Sci. 2009;122(3):301-304.
[26] JIN Z, EL-DEIRY WS. Distinct Signaling Pathways in TRAIL- versus Tumor Necrosis Factor-Induced Apoptosis. Mol Cell Biol. 2006;26(21): 8136-8148.
[27] LEE SY, KOO JM, YOO HS, et al. The expression of TRAIL and its receptors in human osteoarthritic cartilages. Korean J Med.2008;74:296-304.
[28] GOLDRING MB, BERENBAUM F. Emerging targets in osteoarthritis therapy. Curr Opin Pharm. 2015;22:51-63.
[29] 许颖,范凯健,王婷玉.骨关节炎的发病机制及其药物治疗进展[J].实用药物与临床,2018,21(12):104-109.
[30] CHEN D, SHEN J, ZHAO W, et al. Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res. 2017;5:16044.
[31] GEURTS J, PATEL A, HIRSCHMANN MT, et al. Elevated marrow inflammatory cells and osteoclasts in subchondral osteosclerosis in human knee osteoarthritis. J Orthop Res. 2016;34(2):262-269.
[32] 张文贤,王小燕,冯康虎,等.创伤性骨关节炎软骨细胞损坏与修复机制[J].中国组织工程研究,2012,16(46):8727-8732.
[33] SHANG X, WANG Z, TAO H. Mechanism and therapeutic effectiveness of nerve growth factor in osteoarthritis pain. Ther Clin Risk Manag. 2017;13:951-956.
[34] NI Q, TAN Y, ZHANG X, et al. Prenatal ethanol exposure increases osteoarthritis susceptibility in female rat offspring by programming a low-functioning IGF-1 signaling pathway. Sci Rep. 2015;5:14711.
|