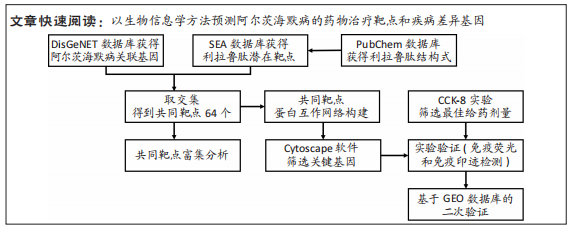
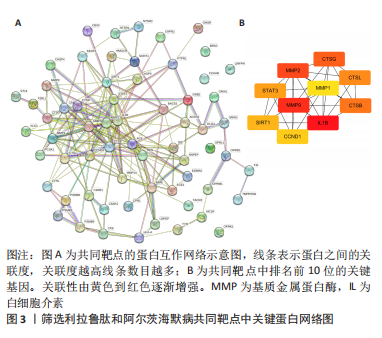
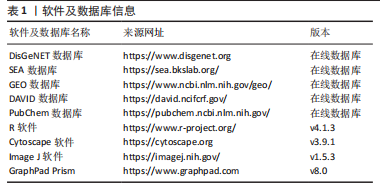
[1] CARMONA S, HARDY J, GUERREIRO R. The genetic landscape of Alzheimer disease. Handb Clin Neurol. 2018;148:395-408.
[2] 2021 Alzheimer’s disease facts and figures. Alzheimers Dement. 2021; 17(3):327-406.
[3] ZHANG Y, LI Y, MA L. Recent advances in research on Alzheimer’s disease in China. J Clin Neurosci. 2020;81:43-46.
[4] KNUDSEN LB, NIELSEN PF, HUUSFELDT PO, et al. Potent derivatives of glucagon-like peptide-1 with pharmacokinetic properties suitable for once daily administration. J Med Chem. 2000;43(9):1664-1669.
[5] NAUCK MA, QUAST DR, WEFERS J, et al. GLP-1 receptor agonists in the treatment of type 2 diabetes - state-of-the-art. Mol Metab. 2021;46:101102.
[6] VILSBOLL T. The effects of glucagon-like peptide-1 on the beta cell. Diabetes Obes Metab. 2009;11 Suppl 3:11-18.
[7] LOVSHIN JA, DRUCKER DJ. Incretin-based therapies for type 2 diabetes mellitus. Nat Rev Endocrinol. 2009;5(5):262-269.
[8] ROLIN B, LARSEN MO, GOTFREDSEN CF, et al. The long-acting GLP-1 derivative NN2211 ameliorates glycemia and increases beta-cell mass in diabetic mice. Am J Physiol Endocrinol Metab. 2002;283(4):E745-E752.
[9] LIU XY, ZHANG N, ZHANG SX, et al. Potential new therapeutic target for Alzheimer’s disease: Glucagon-like peptide-1. Eur J Neurosci. 2021;54(10): 7749-7769.
[10] MENG L, LI XY, SHEN L, et al. Type 2 diabetes mellitus drugs for alzheimer’s disease: current evidence and therapeutic opportunities. Trends Mol Med. 2020;26(6):597-614.
[11] 张峥,张兆辉.阿尔茨海默病与2型糖尿病的相关性研究进展[J].卒中与神经疾病,2018,25(1):112-117.
[12] PALADUGU L, GHARAIBEH A, KOLLI N, et al. Liraglutide has anti-inflammatory and anti-amyloid properties in streptozotocin-induced and 5xFAD mouse models of Alzheimer’s disease. Int J Mol Sci. 2021;22(2):860.
[13] CICCONE L, VANDOOREN J, NENCETTI S, et al. Natural marine and terrestrial compounds as modulators of matrix metalloproteinases-2 (MMP-2) and MMP-9 in Alzheimer’s disease. Pharmaceuticals (Basel). 2021;14(2):86.
[14] REVUELTA M, URRUTIA J, VILLARROEL A, et al. Microglia-mediated inflammation and neural stem cell differentiation in Alzheimer’s disease: possible therapeutic role of K(V)1.3 channel blockade. Front Cell Neurosci. 2022;16:868842.
[15] PY NA, BONNET AE, BERNARD A, et al. Differential spatio-temporal regulation of MMPs in the 5xFAD mouse model of Alzheimer’s disease: evidence for a pro-amyloidogenic role of MT1-MMP. Front Aging Neurosci. 2014;6:247.
[16] HIRANO T, MORI Y. Anti-atherogenic and anti-inflammatory properties of glucagon-like peptide-1, glucose-dependent insulinotropic polypepide, and dipeptidyl peptidase-4 inhibitors in experimental animals. J Diabetes Investig. 2016;7 Suppl 1:80-86.
[17] MCCLEAN PL, GAULT VA, HARRIOTT P, et al. Glucagon-like peptide-1 analogues enhance synaptic plasticity in the brain: a link between diabetes and Alzheimer’s disease. Eur J Pharmacol. 2010;630(1-3):158-162.
[18] HAMILTON A, PATTERSON S, PORTER D, et al. Novel GLP-1 mimetics developed to treat type 2 diabetes promote progenitor cell proliferation in the brain. J Neurosci Res. 2011;89(4):481-489.
[19] QI L, KE L, LIU X, et al. Subcutaneous administration of liraglutide ameliorates learning and memory impairment by modulating tau hyperphosphorylation via the glycogen synthase kinase-3beta pathway in an amyloid beta protein induced alzheimer disease mouse model. Eur J Pharmacol. 2016;783:23-32.
[20] 原丽,韩玲娜,常永丽,等.肠促胰岛素类似物拮抗APP/PS1转基因鼠认知功能损伤的研究[J].中国临床药理学与治疗学,2019,24(7):721-729.
[21] HAN WN, HOLSCHER C, YUAN L, et al. Liraglutide protects against amyloid-beta protein-induced impairment of spatial learning and memory in rats. Neurobiol Aging. 2013;34(2):576-588.
[22] WEINA H, YUHU N, CHRISTIAN H, et al. Liraglutide attenuates the depressive- and anxiety-like behaviour in the corticosterone induced depression model via improving hippocampal neural plasticity. Brain Res. 2018;1694:55-62.
[23] MANNELLO F. New implications of the proteolytic balance between matrix metalloproteinases and their tissue inhibitors in migraine with and without aura. Clin Chim Acta. 2009;409(1-2):1-3.
[24] REMPE RG, HARTZA, BAUER B. Matrix metalloproteinases in the brain and blood-brain barrier: versatile breakers and makers. J Cereb Blood Flow Metab. 2016;36(9):1481-1507.
[25] BJERKE M, ZETTERBERG H, EDMAN A, et al. Cerebrospinal fluid matrix metalloproteinases and tissue inhibitor of metalloproteinases in combination with subcortical and cortical biomarkers in vascular dementia and Alzheimer’s disease. J Alzheimers Dis. 2011;27(3):665-676.
[26] BRUNO MA, MUFSON EJ, WUU J, et al. Increased matrix metalloproteinase 9 activity in mild cognitive impairment. J Neuropathol Exp Neurol. 2009; 68(12):1309-1318.
[27] LORENZL S, BUERGER K, HAMPEL H, et al. Profiles of matrix metalloproteinases and their inhibitors in plasma of patients with dementia. Int Psychogeriatr. 2008;20(1):67-76.
[28] MIZOGUCHI H, TAKUMA K, FUKUZAKI E, et al. Matrix metalloprotease-9 inhibition improves amyloid beta-mediated cognitive impairment and neurotoxicity in mice. J Pharmacol Exp Ther. 2009;331(1):14-22.
[29] WEEKMAN EM, WILCOCK DM. Matrix metalloproteinase in blood-brain barrier breakdown in dementia. J Alzheimers Dis. 2016;49(4):893-903.
[30] ROSENBERG GA. Matrix metalloproteinases and their multiple roles in neurodegenerative diseases. Lancet Neurol. 2009;8(2):205-216.
[31] TEIXEIRA AL, REIS HJ, COELHO FM, et al. All-or-nothing type biphasic cytokine production of human lymphocytes after exposure to Alzheimer’s beta-amyloid peptide. Biol Psychiatry. 2008;64(10):891-895.
[32] HAMPEL H, CARACI F, CUELLO AC, et al. A path toward precision medicine for neuroinflammatory mechanisms in Alzheimer’s disease. Front Immunol. 2020;11:456.
[33] WICINSKI M, SOCHA M, MALINOWSKI B, et al. Liraglutide and its neuroprotective properties-focus on possible biochemical mechanisms in Alzheimer’s disease and cerebral ischemic events. Int J Mol Sci. 2019; 20(5):1050.
[34] KHAVINSON V, LINKOVA N, DYATLOVA A, et al. Senescence-associated secretory phenotype of cardiovascular system cells and inflammaging: perspectives of peptide regulation. Cells. 2022;12(1):106.
[35] SHAFTEL SS, GRIFFIN WS, O’BANION MK. The role of interleukin-1 in neuroinflammation and Alzheimer disease: an evolving perspective. J Neuroinflammation. 2008;5:7.
[36] ZHENG C, ZHOU XW, WANG JZ. The dual roles of cytokines in Alzheimer’s disease: update on interleukins, TNF-alpha, TGF-beta and IFN-gamma. Transl Neurodegener. 2016;5:7. |