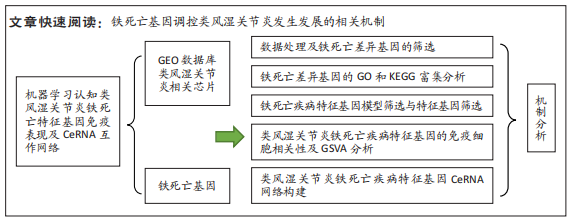
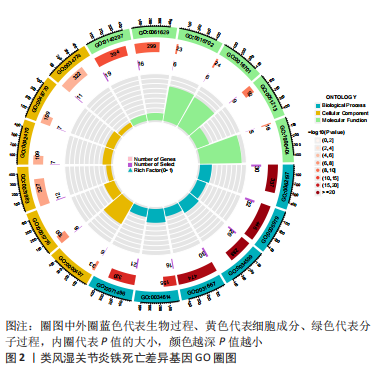
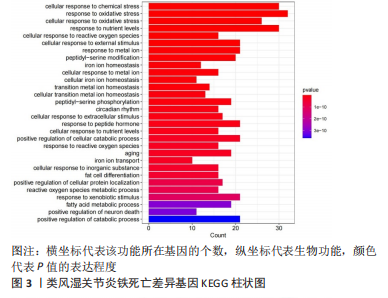
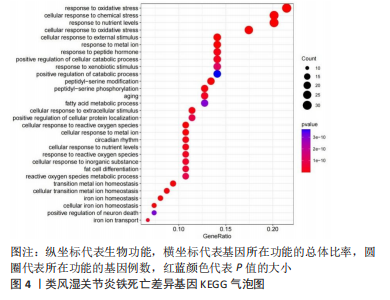
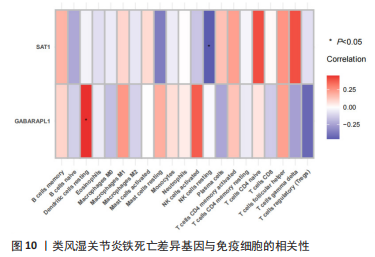
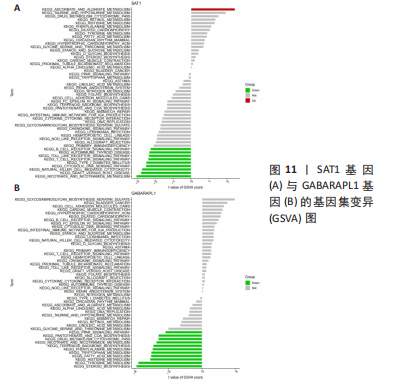
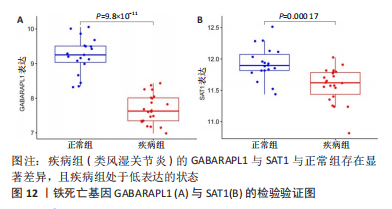
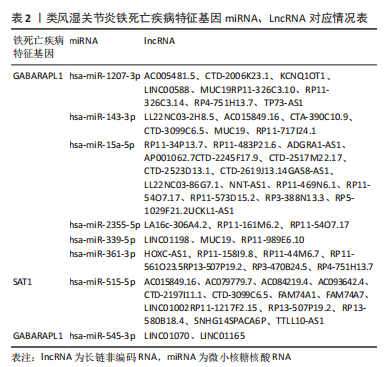
[1] OSIPOVA D, JANSSEN R, MARTENS HA. Rheumatoid arthritis: more than a joint disease. Ned Tijdschr Geneeskd. 2020;164:D4166.
[2] MATEEN S, ZAFAR A, MOIN S, et al. Understanding the role of cytokines in the pathogenesis of rheumatoid arthritis. Clin Chim Acta. 2016;455:161-171.
[3] GIANNINI D, ANTONUCCI M, PETRELLI F, et al. One year in review 2020: pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol. 2020;38(3):387-397.
[4] XIE ZX, HOU HD, LUO D, et al. ROS-Dependent Lipid Peroxidation and Reliant Antioxidant Ferroptosis-Suppressor-Protein 1 in Rheumatoid Arthritis: a Covert Clue for Potential Therapy. Inflammation. 2021;44(1):35-47.
[5] TANSKI W, CHABOWSKI M, JANKOWSKA-POLANSKA B, et al. Iron metabolism in patients with rheumatoid arthritis. Eur Rev Med Pharmacol Sci. 2021; 25(12):4325-4335.
[6] SU LJ, ZHANG JH, GOMEZ H, et al. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxidative Med Cell Longev. 2019;2019:13.
[7] DODSON M, CASTRO-PORTUGUEZ R, ZHANG DD. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019;23:7.
[8] PHULL AR, NASIR B, UL HAQ I, et al. Oxidative stress, consequences and ROS mediated cellular signaling in rheumatoid arthritis. Chem-Biol Interact. 2018;281:121-136.
[9] HUBER R, HUMMERT C, GAUSMANN U, et al. Identification of intra-group, inter-individual, and gene-specific variances in mRNA expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther. 2008;10(4):16.
[10] WOETZEL D, HUBER R, KUPFER P, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther. 2014;16(2):21.
[11] STOCKWELL BR, ANGELI JPF, BAYIR H, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171(2):273-285.
[12] YANG GL, CHANG CC, YANG YW, et al. Resveratrol Alleviates Rheumatoid Arthritis via Reducing ROS and Inflammation, Inhibiting MAPK Signaling Pathways, and Suppressing Angiogenesis. J Agric Food Chem. 2018;66(49): 12953-12960.
[13] FENG FB, QIU HY. Effects of Artesunate on chondrocyte proliferation, apoptosis and autophagy through the PI3K/AKT/mTOR signaling pathway in rat models with rheumatoid arthritis. Biomed Pharmacother. 2018;102:1209-1220.
[14] WOJCIK P, GEGOTEK A, ZARKOVIC N, et al. Oxidative Stress and Lipid Mediators Modulate Immune Cell Functions in Autoimmune Diseases. Int J Mol Sci. 2021;22(2):20.
[15] MAGTANONG L, KO PJ, TO M, et al. Exogenous Monounsaturated Fatty Acids Promote a Ferroptosis-Resistant Cell State. Cell Chem Biol. 2019;26(3):420-432.e9.
[16] DIXON SJ, LEMBERG KM, LAMPRECHT MR, et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell. 2012;149(5):1060-1072.
[17] YANG WS, STOCKWELL BR. Ferrootosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016;26(3):165-176.
[18] GALARIS D, BARBOUTI A, PANTOPOULOS K. Iron homeostasis and oxidative stress: An intimate relationship. Biochim Biophys ActaMol Cell Res. 2019; 1866(12):15.
[19] NAKAMURA T, NAGURO I, ICHIJO H. Iron homeostasis and iron-regulated ROS in cell death, senescence and human diseases. Biochim Biophys ActaGen Subj. 2019;1863(9):1398-1409.
[20] CHOI RY, COYNER AS, KALPATHY-CRAMER J, et al. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl Vis Sci Technol. 2020;9(2):12.
[21] LEE JM, LEE H, KANG S, et al. Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Biotechnological Advances. Nutrients. 2016;8(1):13.
[22] MIRZA AZ, ALTHAGAFI II, SHAMSHAD H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur J Med Chem. 2019;166:502-513.
[23] VENKATESH D, O’BRIEN NA, ZANDKARIMI F, et al. MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev. 2020;34(7-8):526-543.
[24] MARION-LETELLIER R, SAVOYE G, GHOSH S. Fatty acids, eicosanoids and PPAR gamma. Eur J Pharmacol. 2016;785:44-49.
[25] HU ZY, CHEN YD, ZHU SL, et al. Sonic Hedgehog Promotes Proliferation and Migration of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via Rho/ROCK Signaling. J Immunol Res. 2022;2022:9.
[26] ZHU SL, YE YM, SHI YM, et al. Sonic Hedgehog Regulates Proliferation, Migration and Invasion of Synoviocytes in Rheumatoid Arthritis via JNK Signaling. Front Immunol. 2020;11:14.
[27] LIU F, FENG XX, ZHU SL, et al. Sonic Hedgehog Signaling Pathway Mediates Proliferation and Migration of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via MAPK/ERK Signaling Pathway. Front Immunol. 2018;9:10.
[28] WANG MX, ZHU SL, PENG WX, et al. Sonic Hedgehog Signaling Drives Proliferation of Synoviocytes in Rheumatoid Arthritis: A Possible Novel Therapeutic Target. J Immunol Res. 2014;2014:10.
[29] HUANG SS, CHENG H, TANG CM, et al. Anti-oxidative, anti-apoptotic, and pro-angiogenic effects mediate functional improvement by sonic hedgehog against focal cerebral ischemia in rats. Exp Neurol. 2013;247:680-688.
[30] ZHOU L, HUANG LT, XU QL, et al. Association of MUC19 Mutation With Clinical Benefits of Anti-PD-1 Inhibitors in Non-small Cell Lung Cancer. Front Oncol. 2021;11:12.
[31] KERSCHNER JE, KHAMPANG P, ERBE CB, et al. Mucin gene 19 (MUC19) expression and response to inflammatory cytokines in middle ear epithelium. Glycoconjugate J. 2009;26(9):1275-1284.
[32] TANG ZY, ZENG XH, LI JJ, et al. LncRNA HOXC-AS1 promotes nasopharyngeal carcinoma (NPC) progression by sponging miR-4651 and subsequently upregulating FOXO6. J Pharmacol Sci. 2021;147(3):284-293.
[33] YU S, YU M, HE X, et al. KCNQ1OT1 promotes autophagy by regulating miR-200a/FOXO3/ATG7 pathway in cerebral ischemic stroke. Aging Cell. 2019;18(3):e12940.
[34] ZHUANG SW, MA Y, ZENG YX, et al. METTL14 promotes doxorubicin-induced cardiomyocyte ferroptosis by regulating the KCNQ1OT1-miR-7-5p-TFRC axis. Cell Biol Toxicol. 2021. doi: 10.1007/s10565-021-09660-7.
|