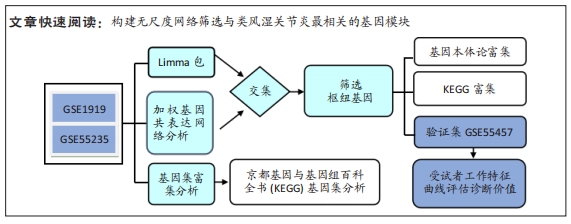
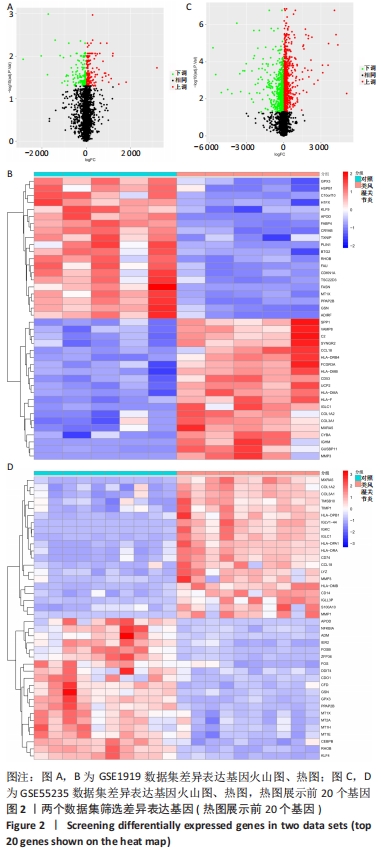
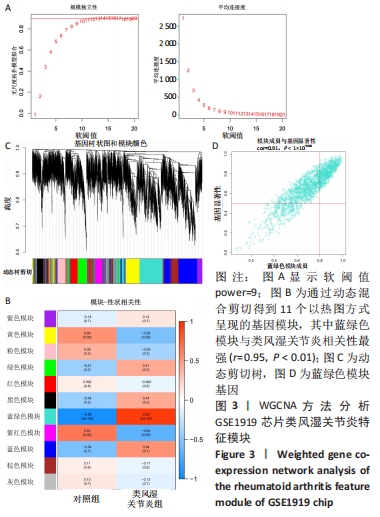
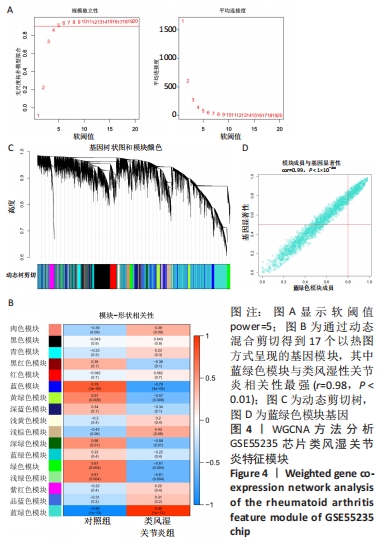
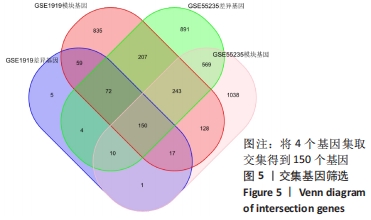
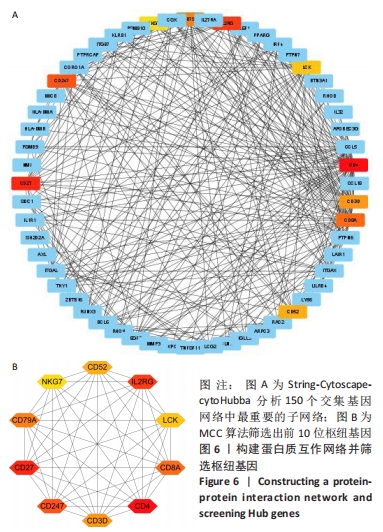
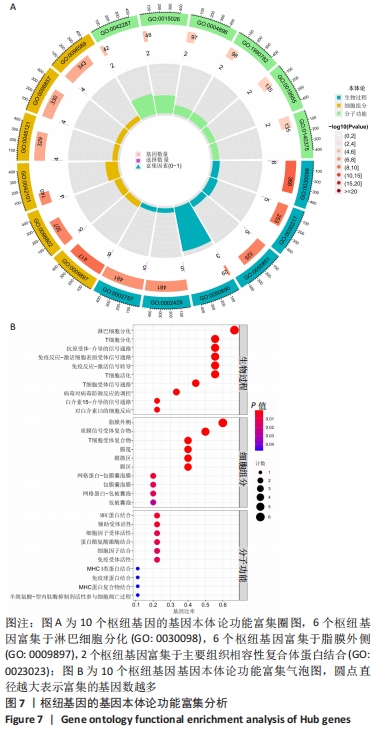
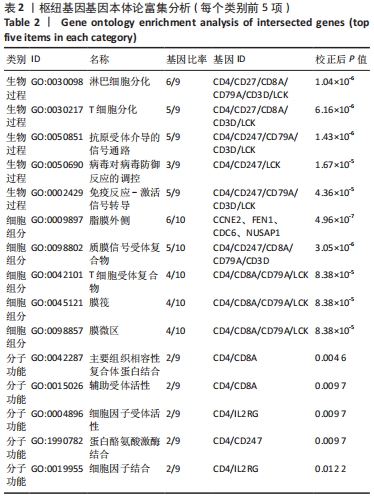
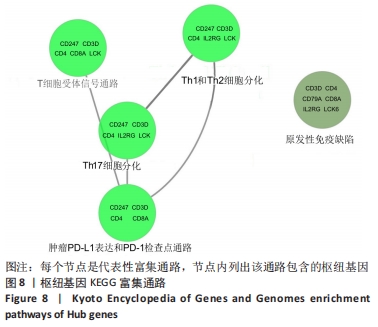
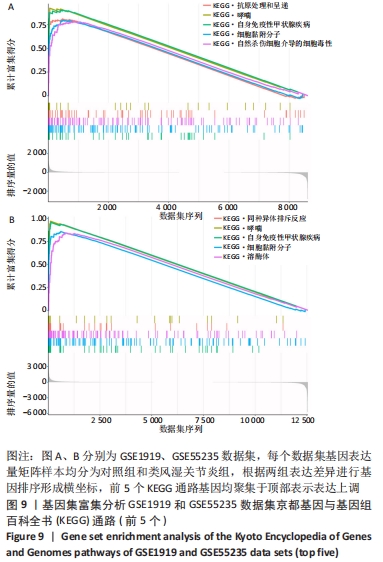
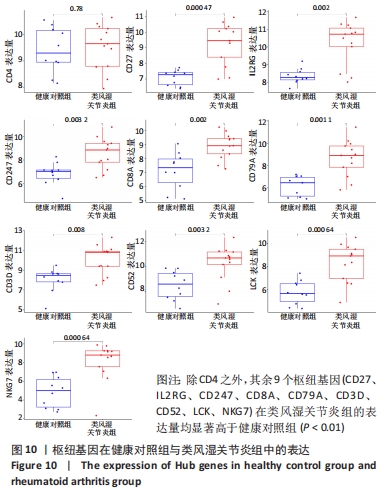
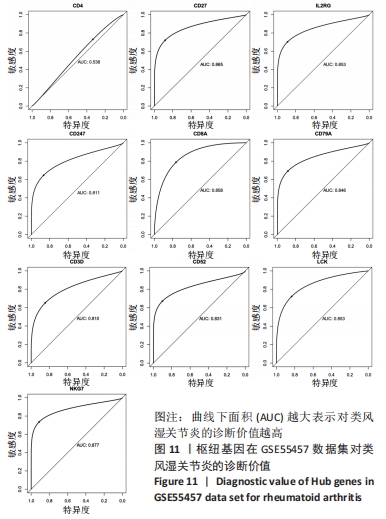
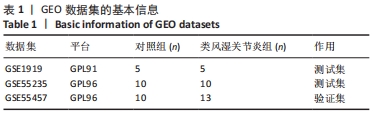
[1] 周新朋,金晔华,张润润,等.类风湿关节炎成纤维样滑膜细胞lncRNA-miRNA-mRNA网络构建及关键基因预测[J]. 中国医药导报, 2021,18(9):12-16.
[2] ANAPARTI V, AGARWAL P, SMOLIK I, et al. Whole Blood Targeted Bisulfite Sequencing and Differential Methylation in the C6ORF10 Gene of Patients with Rheumatoid Arthritis. J Rheumatol. 2020;47(11):1614-1623.
[3] ZHOU S, LU H, XIONG M. Identifying Immune Cell Infiltration and Effective Diagnostic Biomarkers in Rheumatoid Arthritis by Bioinformatics Analysis. Front Immunol. 2021;12:1-12.
[4] LONG NP, PARK S, ANH NH, et al. Efficacy of Integrating a Novel 16-Gene Biomarker Panel and Intelligence Classifiers for Differential Diagnosis of Rheumatoid Arthritis and Osteoarthritis. J Clin Med. 2019;8(1):1-16.
[5] KMIOLEK T, PARADOWSKA-GORYCKA A. miRNAs as Biomarkers and Possible Therapeutic Strategies in Rheumatoid Arthritis. Cells. 2022; 11(3):1-22.
[6] GOMES DA SILVA IIF, BARBOSA AD, SOUTO FO, et al. MYD88, IRAK3 and Rheumatoid Arthritis pathogenesis: Analysis of differential gene expression in CD14 + monocytes and the inflammatory cytokine levels. Immunobiology. 2021;226(6):1-16.
[7] HUA L, ZHOU P, LIU H, et al. Mining susceptibility gene modules and disease risk genes from SNP data by combining network topological properties with support vector regression. J Theor Biol. 2011;289:225-236.
[8] 史晋宇,乔军,胡方媛. 基于基因芯片公共数据库及生物信息学方法筛选类风湿关节炎滑膜组织关键基因及通路[J]. 中华风湿病学杂志,2020,24(11):743-746.
[9] WU R, LONG L, ZHOU Q, et al. Identification of hub genes in rheumatoid arthritis through an integrated bioinformatics approach. J Orthop Surg Res. 2021;16(1):458-466.
[10] HU H, ZHANG F, LI L, et al. Identification and Validation of ATF3 Serving as a Potential Biomarker and Correlating With Pharmacotherapy Response and Immune Infiltration Characteristics in Rheumatoid Arthritis. Front Mol Biosci. 2021;8:1-18.
[11] MA C, LV Q, TENG S, et al. Identifying key genes in rheumatoid arthritis by weighted gene co-expression network analysis. Int J Rheum Dis. 2017;20(8):971-979.
[12] REN C, LI M, ZHENG Y, et al. Identification of diagnostic genes and vital microRNAs involved in rheumatoid arthritis: based on data mining and experimental verification. Peer J. 2021;9:1-24.
[13] HOU J, YE X, LI C, et al. K-Module Algorithm: An Additional Step to Improve the Clustering Results of WGCNA Co-Expression Networks. Genes (Basel). 2021;12(1):1-15.
[14] HE P, MO XB, LEI SF, et al. Epigenetically regulated co-expression network of genes significant for rheumatoid arthritis. Epigenomics. 2019;11(14):1-12.
[15] REN C, LI M, DU W, et al. Comprehensive Bioinformatics Analysis Reveals Hub Genes and Inflammation State of Rheumatoid Arthritis. Biomed Res Int. 2020;2020:1-13.
[16] CHEN Y, LIAO R, YAO Y, et al. Machine learning to identify immune-related biomarkers of rheumatoid arthritis based on WGCNA network. Clin Rheumatol. 2021;41(4):1057-1068.
[17] CAI P, JIANG T, LI B, et al. Comparison of rheumatoid arthritis (RA) and osteoarthritis (OA) based on microarray profiles of human joint fibroblast-like synoviocytes. Cell Biochem Funct. 2019;37(1):31-41.
[18] WOODWARD AA, TAYLOR DM, GOLDMUNTZ E, et al. Gene-Interaction-Sensitive enrichment analysis in congenital heart disease. BioData Min. 2022;15(1):1-16.
[19] LU J, BI Y, ZHU Y, et al. CD3D, GZMK, and KLRB1 Are Potential Markers for Early Diagnosis of Rheumatoid Arthritis, Especially in Anti-Citrullinated Protein Antibody-Negative Patients. Front Pharmacol. 2021;12:1-12.
[20] WENJING Z, YANG C, QIN L, et al. Emerging nanotherapeutics alleviating rheumatoid arthritis by readjusting the seeds and soils. J Control Release. 2022;345:851-879.
[21] FERNANDA LF, BRUNO N, EDSON M, et al. Rheumatoid Meningitis as an Extra-articular Manifestation of Rheumatoid Arthritis. Neurol India. 2021;69(6):1903-1904.
[22] ZECHEN M, HANYAN Z, MARY-CLAIR Y, et al. Prevalence and trajectory of erosions, synovitis, and bone marrow edema in feet of patients with early rheumatoid arthritis. Clin Rheumatol. 2021;40(9):3575-3579.
[23] CHAOJUN W, YONG Q, JINGHUA X, et al. Correlation of Ultrasound Synovitis Joint Count with Disease Activity and Its Longitudinal Variation with Treatment Response to Etanercept in Rheumatoid Arthritis. Ultrasound Med Biol. 2021;47(9):2543-2549.
[24] OMA I, OLSTAD OK, ANDERSEN JK, et al. Differential expression of vitamin D associated genes in the aorta of coronary artery disease patients with and without rheumatoid arthritis. PLoS One. 2018;13(8): 1-13.
[25] JABADO O, MALDONADO MA, SCHIFF M, et al. Differential Changes in ACPA Fine Specificity and Gene Expression in a Randomized Trial of Abatacept and Adalimumab in Rheumatoid Arthritis. Rheumatol Ther. 2022;9(2):391-409.
[26] QIAN K, ZHENG XX, WANG C, et al. β-Sitosterol Inhibits Rheumatoid Synovial Angiogenesis Through Suppressing VEGF Signaling Pathway. Front Pharmacol. 2022;12:1-13.
[27] ELLIOTT SE, KONGPACHITH S, LINGAMPALLI N, et al. Affinity Maturation Drives Epitope Spreading and Generation of Proinflammatory Anti-Citrullinated Protein Antibodies in Rheumatoid Arthritis. Arthritis Rheumatol. 2018;70(12):1946-1958.
[28] NAGAI K, ISHII T, OHNO T, et al. Overload of the Temporomandibular Joints Accumulates gammadelta T Cells in a Mouse Model of Rheumatoid Arthritis: A Morphological and Histological Evaluation. Front Immunol. 2021;12(753754):1-10.
[29] XIAO J, WANG R, CAI X, et al. Coupling of Co-expression Network Analysis and Machine Learning Validation Unearthed Potential Key Genes Involved in Rheumatoid Arthritis. Front Genet. 2021;12:1-19.
[30] NOVALIA P, THIBAULT H, LIYAN YS, et al. Induction of antigen-specific tolerance by nanobody-antigen adducts that target class-II major histocompatibility complexes. Nat Biomed Eng. 2021;5(11):1389-1401.
[31] ANIRUDDHA B, PARASAR G, ALAKENDU G, et al. Role of oxidative stress in induction of trans-differentiation of neutrophils in patients with rheumatoid arthritis. Free Radic Res. 2022;22:1-13.
[32] LI WC, BAI L, XU Y, et al. Identification of differentially expressed genes in synovial tissue of rheumatoid arthritis and osteoarthritis in patients. J Cell Biochem. 2019;120(3):4533-4544.
[33] HUSSIEN Y, DAMIATI L, ALHARBI A, et al. Correlations of TGF-β1 (869C/T), CD4- (11743 A/C) and CD4- (10845 A/G) Polymorphism with Biochemical Risk Factor Predict in Rheumatoid Arthritis Patients Progression. Annals of the Rheumatic Diseases. 2015;74(Suppl 2): 987.981-987.
[34] ZHANG R, YANG X, WANG J, et al. Identification of potential biomarkers for differential diagnosis between rheumatoid arthritis and osteoarthritis via integrative genomewide gene expression profiling analysis. Mol Med Rep. 2019;19(1):30-40.
[35] HU F, ZHANG W, SHI L, et al. Impaired CD27(+)IgD(+) B Cells With Altered Gene Signature in Rheumatoid Arthritis. Front Immunol. 2018; 9:626-635.
[36] YE Y, LIU M, TANG L, et al. Iguratimod represses B cell terminal differentiation linked with the inhibition of PKC/EGR1 axis. Arthritis Res Ther. 2019;21(1):1-14.
[37] ABREU M, CARVALHEIRO H, DUARTE C, et al. Expression of CD62L, CD27 and CD11b define a marked cytotoxic effector role for NK cells in rheumatoid arthritis. Ann Rheum Dis. 2011;70(Suppl 2):1-94.
[38] BURSKA AN, THU A, PARMAR R, et al. Quantifying circulating Th17 cells by qPCR: potential as diagnostic biomarker for rheumatoid arthritis. Rheumatology (Oxford). 2019;58(11):2015-2024.
|