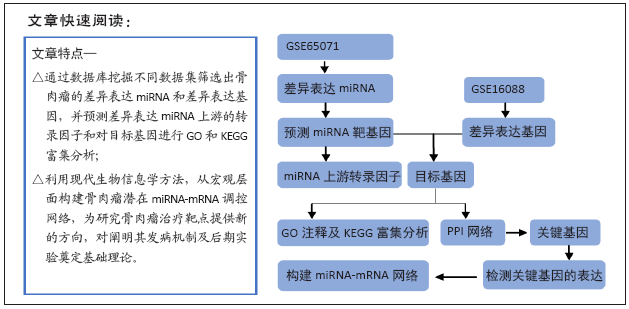
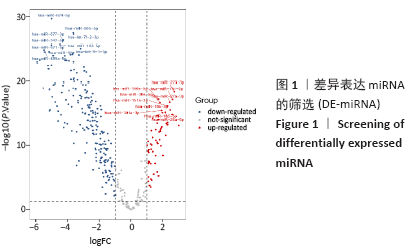
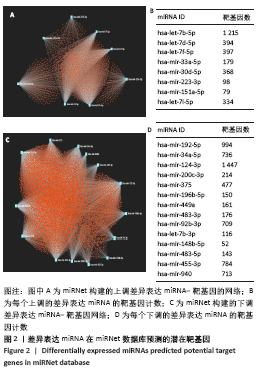
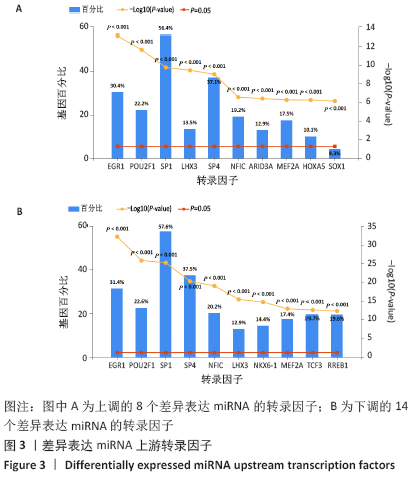
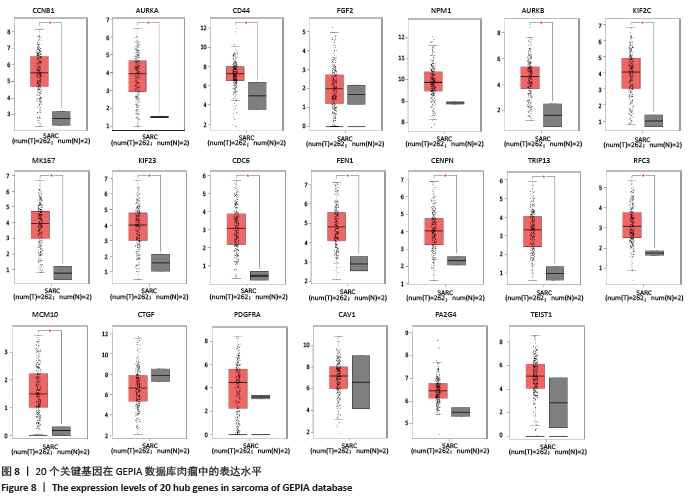
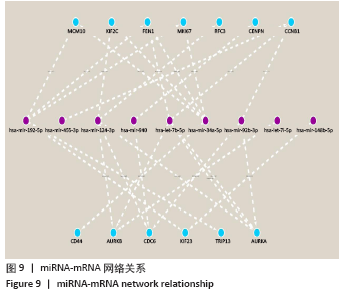
[1] PERUT F, RONCUZZI L, BALDINI N. The Emerging Roles of Extracellular Vesicles in Osteosarcoma. Front Oncol. 2019;9:1342.
[2] ABU-HALIMA M, BACKES C, LEIDINGER P, et al. MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil Steril. 2014;101(1):78-86.e2.
[3] HUERTA-ZAVALA ML, LOPEZ-CASTILLEJOS ES, REQUENEZ-CONTRERAS JL, et al. A single miRNA and miRNA sponge expression system for efficient modulation of miR-223 availability in mammalian cells. J Gene Med. 2019; 21(8):e3100.
[4] ANDERSEN GB, KNUDSEN A, HAGER H, et al. miRNA profiling identifies deregulated miRNAs associated with osteosarcoma development and time to metastasis in two large cohorts. Mol Oncol. 2018;12(1):114-131.
[5] GONG N, GONG M. MiRNA-221 from tissue may predict the prognosis of patients with osteosarcoma. Medicine (Baltimore). 2018;97(29):e11100.
[6] 张琨. miR-125b, miR-126, miR-145在骨肉瘤病人血清中的表达[D].长沙:中南大学,2014.
[7] 陈康.骨肉瘤病人血清中miRNA的表达谱研究[D]. 南京:南京大学, 2012.
[8] 刘书中,劳立峰,王以朋.外周血miRNAs检测在骨肉瘤中的意义[J].中国生物制品学杂志,2016,29(5):552-556.
[9] PLOTNIKOVA OM, SKOBLOV MY. Efficiency of the miRNA- mRNA Interaction Prediction Programs. Mol Biol (Mosk). 2018;52(3):543-554.
[10] BHOWMICK SS, BHATTACHARJEE D, RATO L. Integrated analysis of the miRNA-mRNA next-generation sequencing data for finding their associations in different cancer types. Comput Biol Chem. 2020;84:107152.
[11] SHEKHAR R, PRIYANKA P, KUMAR P, et al. The microRNAs miR-449a and miR-424 suppress osteosarcoma by targeting cyclin A2 expression. J Biol Chem. 2019;294(12):4381-4400.
[12] LIU G, HUANG K, JIE Z, et al. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol Cancer. 2018;17(1):170.
[13] WANG Y, ZHANG S, XU Y, et al. Upregulation of miR-192 inhibits cell growth and invasion and induces cell apoptosis by targeting TCF7 in human osteosarcoma. Tumour Biol. 2016;37(11):15211-15220.
[14] 隆姝孜,周寒静,刘静姝,等. mir-151a-5p对肺癌A549细胞生物学功能和化疗敏感性的影响[J].第三军医大学学报,2019,41(8):770-777.
[15] LONG HD, MA YS, YANG HQ, et al. Reduced hsa-miR-124-3p levels are associated with the poor survival of patients with hepatocellular carcinoma. Mol Biol Rep. 2018;45(6):2615-2623.
[16] LI S, WU X, PEI Y, et al. PTHR1 May Be Involved in Progression of Osteosarcoma by Regulating miR-124-3p-AR-Tgfb1i1, miR-27a-3p-PPARG-Abca1, and miR-103/590-3p-AXIN2 Axes. DNA Cell Biol. 2019;38(11):1323-1337.
[17] JI Q, XU X, SONG Q, et al. miR-223-3p Inhibits Human Osteosarcoma Metastasis and Progression by Directly Targeting CDH6. Mol Ther. 2018; 26(5):1299-1312.
[18] MULLANY LE, HERRICK JS, WOLFF RK, et al. MicroRNA-transcription factor interactions and their combined effect on target gene expression in colon cancer cases. Version 2. Genes Chromosomes Cancer. 2018;57(4):192-202.
[19] DHARAP A, POKRZYWA C, MURALI S, et al. Mutual induction of transcription factor PPARγ and microRNAs miR-145 and miR-329. J Neurochem. 2015; 135(1):139-146.
[20] YANG W, LU Z, ZHI Z, et al. Increased miRNA-518b inhibits trophoblast migration and angiogenesis by targeting EGR1 in early embryonic arrest. Biol Reprod. 2019;101(4):664-674.
[21] 秦丽,张惠华,郭淑军,等.外源性EGR1瞬时表达对骨肉瘤细胞细胞周期和凋亡的影响[J].中国病理生理杂志,2011,27(2):293-299.
[22] NG MC, LAM VK, TAM CH, et al. Association of the POU class 2 homeobox 1 gene (POU2F1) with susceptibility to Type 2 diabetes in Chinese populations. Diabet Med. 2010;27(12):1443-1449.
[23] STEPCHENKO AG, GEORGIEVA SG, PANKRATOVA EV. Multiple Interactions of the Oct-1 (POU2F1) Transcription Factor with PORE and MORE Sites. Mol Biol (Mosk). 2019;53(3):430-435.
[24] VÁZQUEZ-ARREGUÍN K, TANTIN D. The Oct1 transcription factor and epithelial malignancies: Old protein learns new tricks. Biochim Biophys Acta. 2016;1859(6):792-804.
[25] XIE CH, CAO YM, HUANG Y, et al. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F1 expression. Tumour Biol. 2016;37(11):15031-15041.
[26] PENG F, ZHOU Y, WANG J, et al. The transcription factor Sp1 modulates RNA polymerase III gene transcription by controlling BRF1 and GTF3C2 expression in human cells. J Biol Chem. 2020;295(14):4617-4630.
[27] MENG Y, HAO D, HUANG Y, et al. Positive feedback loop SP1/MIR17HG/miR-130a-3p promotes osteosarcoma proliferation and cisplatin resistance. Biochem Biophys Res Commun. 2020;521(3):739-745.
[28] HOU Z, GUO K, SUN X, et al. TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling. Mol Cancer. 2018;17(1):172.
[29] HE L, XU J, CHEN L, et al. Apelin/APJ signaling in hypoxia-related diseases. Clin Chim Acta. 2015;451(Pt B):191-198.
[30] CHENG B, CHEN J, BAI B, et al. Neuroprotection of apelin and its signaling pathway. Peptides. 2012;37(1):171-173.
[31] 郝延璋. Apelin对胃癌生物学功能的影响及其分子机制的研究[D].济南:山东大学,2017.
[32] CHEN T, LIU N, XU GM, et al. Apelin13/APJ promotes proliferation of colon carcinoma by activating Notch3 signaling pathway. Oncotarget. 2017;8(60): 101697-101706.
[33] THEOCHARIS AD, KARAMANOS NK. Proteoglycans remodeling in cancer: Underlying molecular mechanisms. Matrix Biol. 2019;75-76:220-259.
[34] YE C, WANG J, WU P, et al. Prognostic role of cyclin B1 in solid tumors: a meta-analysis. Oncotarget. 2017;8(2):2224-2232.
[35] ZHANG J, ZHU X, LI H, et al. Piperine inhibits proliferation of human osteosarcoma cells via G2/M phase arrest and metastasis by suppressing MMP-2/-9 expression. Int Immunopharmacol. 2015;24(1):50-58.
[36] VADER G, LENS SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786(1):60-72.
[37] D’ASSORO AB, HADDAD T, GALANIS E. Aurora-A Kinase as a Promising Therapeutic Target in Cancer. Front Oncol. 2016;5:295.
[38] JIANG Z, JIANG J, YANG H, et al. Silencing of Aurora kinase A by RNA interference inhibits tumor growth in human osteosarcoma cells by inducing apoptosis and G2/M cell cycle arrest. Oncol Rep. 2014;31(3):1249-1254.
[39] CICHY J, PURÉ E. The liberation of CD44. J Cell Biol. 2003;161(5):839-843.
[40] PONTA H, SHERMAN L, HERRLICH PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33-45.
[41] SKANDALIS SS, GIALELI C, THEOCHARIS AD, et al. Advances and advantages of nanomedicine in the pharmacological targeting of hyaluronan-CD44 interactions and signaling in cancer. Adv Cancer Res. 2014;123:277-317.
[42] XIAO Z, WAN J, NUR AA, et al. Targeting CD44 by CRISPR-Cas9 in Multi-Drug Resistant Osteosarcoma Cells. Cell Physiol Biochem. 2018;51(4):1879-1893.
|