[1] LI R, LIN QX, LIANG XZ, et al. Stem cell therapy for treating osteonecrosis of the femoral head: from clinical applications to related basic research. Stem Cell Res Ther. 2018;9(1):291.
[2] 孙笛,赵李奔,刘春晖,等.股骨头坏死危险因素流行病学分析[J].中华肿瘤防治杂志,2016,23(S1):10-11.
[3] ZHAO DW, YU M, HU K, et al. Prevalence of nontraumatic osteonecrosis of the femoral head and its associated risk factors in the chinese population: results from a nationally representative survey. Chin Med J (Engl). 2015;128(21):2843-2850.
[4] 曾祥洪,梁博伟.股骨头坏死保髋治疗的新策略[J].中国组织工程研究,2021,25(3):431-437.
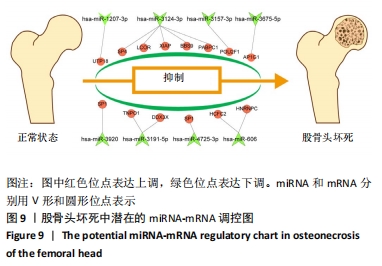
[5] 张颖,张蕾蕾,柴玉娜,等.创伤性股骨头坏死中差异miRNA的筛选与验证[J].中华创伤骨科杂志,2017,19(11):978-985.
[6] GENNARI L, BIANCIARDI S, MERLOTTI D. MicroRNAs in bone diseases. Osteoporos Int. 2017;28(4):1191-1213.
[7] 中国医师协会骨科医师分会骨循环与骨坏死专业委员会,中华医学会骨科分会骨显微修复学组,国际骨循环学会.中国区中国成人股骨头坏死临床诊疗指南(2020)[J].中华骨科杂志,2020,40(20): 1365-1376.
[8] CUI D, ZHAO D, WANG B, et al. Safflower (Carthamus tinctorius L.) polysaccharide attenuates cellular apoptosis in steroid-induced avascular necrosis of femoral head by targeting caspase-3-dependent signaling pathway. Int J Biol Macromol. 2018;116:106-112.
[9] ZHANG Y, YIN J, DING H, et al. Vitamin k2 ameliorates damage of blood vessels by glucocorticoid: a potential mechanism for its protective effects in glucocorticoid-induced osteonecrosis of the femoral head in a rat model. Int J Biol Sci. 2016;12(7):776-785.
[10] LI GQ, WANG ZY. MiR-20b promotes osteocyte apoptosis in rats with steroid-induced necrosis of the femoral head through BMP signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(11):4599-4608.
[11] DONG Y, LI T, LI Y, et al. MicroRNA-23a-3p inhibitor decreases osteonecrosis incidence in a rat model. Mol Med Rep. 2017;16(6): 9331-9336.
[12] LIU Y, ZONG Y, SHAN H, et al. MicroRNA-23b-3p participates in steroid-induced osteonecrosis of the femoral head by suppressing ZNF667 expression. Steroids. 2020;163:108709.
[13] XU W, LI J, TIAN H, et al. MicroRNA‑186‑5p mediates osteoblastic differentiation and cell viability by targeting CXCL13 in non‑traumatic osteonecrosis. Mol Med Rep. 2019;20(5):4594-4602.
[14] WANG C, SUN W, LING S, et al. AAV-Anti-miR-214 prevents collapse of the femoral head in osteonecrosis by regulating osteoblast and osteoclast activities. Mol Ther Nucleic Acids. 2019;18:841-850.
[15] FANG SH, CHEN L, CHEN HH, et al. MiR-15b ameliorates SONFH by targeting Smad7 and inhibiting osteogenic differentiation of BMSCs. Eur Rev Med Pharmacol Sci. 2019;23(22):9761-9771.
[16] YU L, XU Y, QU H, et al. Decrease of MiR-31 induced by TNF-α inhibitor activates SATB2/RUNX2 pathway and promotes osteogenic differentiation in ethanol-induced osteonecrosis. J Cell Physiol. 2019; 234(4):4314-4326.
[17] ZHANG Y, WEI QS, DING WB, et al. Increased microRNA-93-5p inhibits osteogenic differentiation by targeting bone morphogenetic protein-2. PLoS One. 2017;12(8):e0182678.
[18] LIAO W, NING Y, XU HJ, et al. BMSC-derived exosomes carrying microRNA-122-5p promote proliferation of osteoblasts in osteonecrosis of the femoral head. Clin Sci (Lond). 2019;133(18):1955-1975.
[19] SUN Z, WU F, YANG Y, et al. MiR-144-3p Inhibits BMSC Proliferation and Osteogenic Differentiation Via Targeting FZD4 in Steroid-Associated Osteonecrosis. Curr Pharm Des. 2019;25(45):4806-4812.
[20] ZHANG WL, CHI CT, MENG XH. miRNA‑15a‑5p facilitates the bone marrow stem cell apoptosis of femoral head necrosis through the Wnt/β‑catenin/PPARγ signaling pathway. Mol Med Rep. 2019;19(6): 4779-4787.
[21] MENG CY, XUE F, ZHAO ZQ, et al. Influence of MicroRNA-141 on Inhibition of the proliferation of bone marrow mesenchymal stem cells in steroid-induced osteonecrosis via SOX11. Orthop Surg. 2020; 12(1):277-285.
[22] XIE Y, HU JZ, SHI ZY. MiR-181d promotes steroid-induced osteonecrosis of the femoral head by targeting SMAD3 to inhibit osteogenic differentiation of hBMSCs. Eur Rev Med Pharmacol Sci. 2018;22(13):4053-4062.
[23] XU HJ, LIAO W, LIU XZ, et al. Down-regulation of exosomal microRNA-224-3p derived from bone marrow-derived mesenchymal stem cells potentiates angiogenesis in traumatic osteonecrosis of the femoral head. FASEB J. 2019;33(7):8055-8068.
[24] TIAN ZJ, LIU BY, ZHANG YT, et al. MiR-145 silencing promotes steroid-induced avascular necrosis of the femoral head repair via upregulating VEGF. Eur Rev Med Pharmacol Sci. 2017;21(17):3763-3769.
[25] PENG WX, YE C, DONG WT, et al. MicroRNA-34a alleviates steroid-induced avascular necrosis of femoral head by targeting Tgif2 through OPG/RANK/RANKL signaling pathway. Exp Biol Med (Maywood). 2017; 242(12):1234-1243.
[26] ZHA X, SUN B, ZHANG R, et al. Regulatory effect of microRNA-34a on osteogenesis and angiogenesis in glucocorticoid-induced osteonecrosis of the femoral head. J Orthop Res. 2018;36(1):417-424.
[27] KONG L, ZUO R, WANG M, et al. Silencing microRNA-137-3p, which targets RUNX2 and CXCL12 prevents steroid-induced osteonecrosis of the femoral head by facilitating osteogenesis and angiogenesis. Int J Biol Sci. 2020;16(4):655-670.
[28] CHAO PC, CUI MY, LI XA, et al. Correlation between miR-1207-5p expression with steroid-induced necrosis of femoral head and VEGF expression. Eur Rev Med Pharmacol Sci. 2019;23(7):2710-2718.
[29] LIU XD, CAI F, LIU L, et al. MicroRNA-210 is involved in the regulation of postmenopausal osteoporosis through promotion of VEGF expression and osteoblast differentiation. J Biol Chem. 2015;396(4):339-347.
[30] GU C, XU Y, ZHANG S, et al. miR-27a attenuates adipogenesis and promotes osteogenesis in steroid-induced rat BMSCs by targeting PPARγ and GREM1. Sci Rep. 2016;6:38491.
[31] SUN J, WANG Y, LI Y, et al. Downregulation of PPARγ by miR-548d-5p suppresses the adipogenic differentiation of human bone marrow mesenchymal stem cells and enhances their osteogenic potential. J Transl Med. 2014;12:168.
[32] ZHAO SR, WEN JJ, MU HB. Role of Hsa-miR-122-3p in steroid-induced necrosis of femoral head. Eur Rev Med Pharmacol Sci. 2019;23(3 Suppl): 54-59.
[33] LI G, LIU H, ZHANG X, et al. The protective effects of microRNA-26a in steroid-induced osteonecrosis of the femoral head by repressing EZH2. Cell Cycle. 2020;19(5):551-566.
[34] WEI JH, LUO QQ, TANG YJ, et al. Upregulation of microRNA-320 decreases the risk of developing steroid-induced avascular necrosis of femoral head by inhibiting CYP1A2 both in vivo and in vitro. Gene. 2018;660:136-144.
[35] HUANG S, LI Y, WU P, et al. microRNA-148a-3p in extracellular vesicles derived from bone marrow mesenchymal stem cells suppresses SMURF1 to prevent osteonecrosis of femoral head. J Cell Mol Med. 2020;24(19):11512-11523.
[36] 董政权,魏垒.骨关节炎基因差异谱的生物信息学分析[J].中国组织工程研究,2019,23(3):335-340.
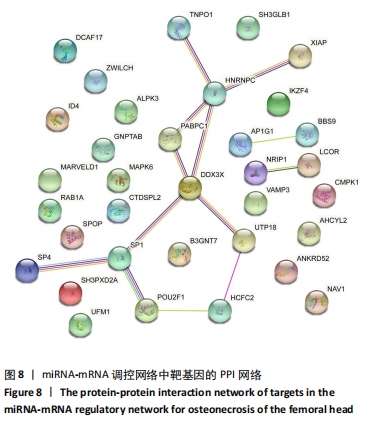
[37] SAFE S, IMANIRAD P, SREEVALSAN S, et al. Transcription factor Sp1, also known as specificity protein 1 as a therapeutic target. Expert Opin Ther Targets. 2014;18(7):759-769.
[38] LIU D, WANG Y, PAN Z, et al. cAMP regulates 11β-hydroxysteroid dehydrogenase-2 and Sp1 expression in MLO-Y4/MC3T3-E1 cells. Exp Ther Med. 2020;20(3):2166-2172.
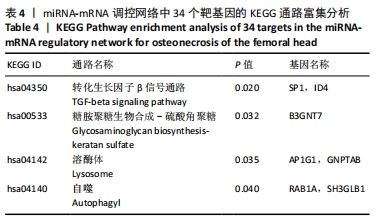
[39] 许灿宏,陈跃平,章晓云.成骨信号通路在非创伤性股骨头坏死中的作用[J].中国组织工程研究,2020,24(14):2235-2242.
[40] TAO J, DONG B, YANG LX, et al. TGF‑β1 expression in adults with non‑traumatic osteonecrosis of the femoral head. Mol Med Rep. 2017; 16(6):9539-9544.
[41] BAI Y, LIU Y, JIN S, et al. Expression of microRNA‑27a in a rat model of osteonecrosis of the femoral head and its association with TGF‑β/Smad7 signalling in osteoblasts. Int J Mol Med. 2019;43(2):850-860.
|