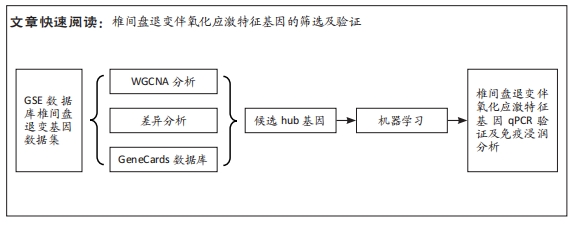
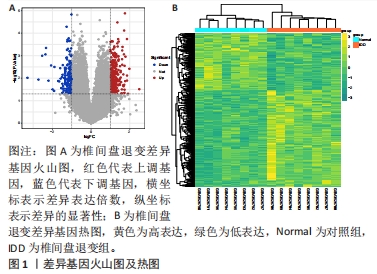
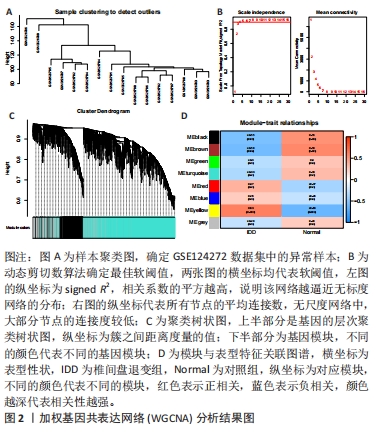
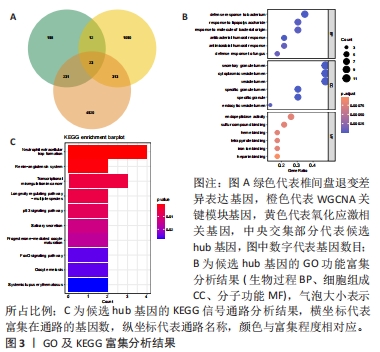
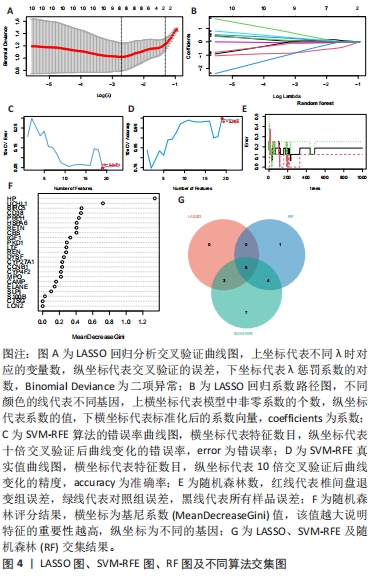
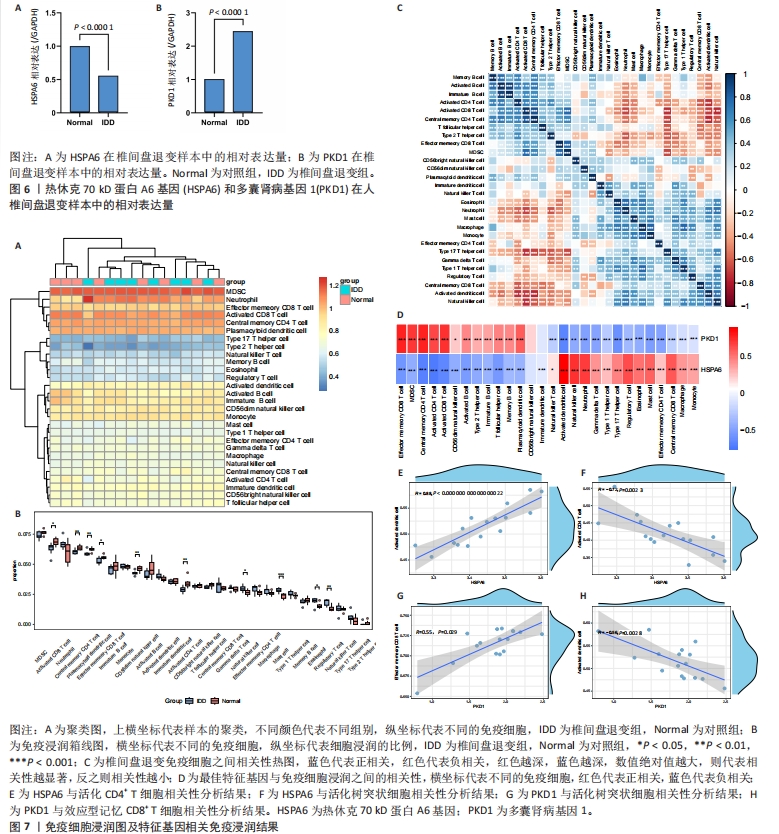
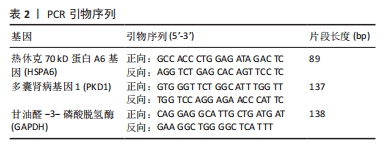
[1] MOHD II, TEOH SL, MOHD NN, et al. Discogenic low back pain: anatomy, pathophysiology and treatments of intervertebral disc degeneration. Int J Mol Sci. 2022;24(1):208-224.
[2] DIWAN AD, MELROSE J. Intervertebral disc degeneration and how it leads to low back pain. JOR Spine. 2023;6(1):e1231.
[3] 史文生.MRI对下腰痛者椎间盘退变的评价价值[J].影像研究与医学应用,2023,7(21):57-59.
[4] FRANCISCO V, PINO J, GONZÁLEZ-GAY MÁ, et al. A new immunometabolic perspective of intervertebral disc degeneration. Nat Rev Rheumatol. 2022; 18(1):47-60.
[5] ROH EJ, DARAI A, KYUNG JW, et al. Genetic therapy for intervertebral disc degeneration. Int J Mol Sci. 2021;22(4):1579-1593.
[6] CHENG F, YANG H, CHENG Y, et al. The role of oxidative stress in intervertebral disc cellular senescence. Front Endocrinol (Lausanne). 2022; 13(6):1038171.
[7] ZHAO Y, XIANG Q, LIN J, et al. Oxidative stress in intervertebral disc degeneration: new insights from bioinformatic strategies. Oxid Med Cell Longev. 2022;2022:2239770.
[8] 周明瀚,张慧,郑先波,等.不同氧浓度下髓核细胞中PI3K/Akt/HIF-1α信号通路表达对延缓椎间盘退变的意义[J].中国组织工程研究,2024, 28(28):4491-4497.
[9] HUANG X, LIU S, WU L, et al. High Throughput single cell RNA sequencing, bioinformatics analysis and applications. Adv Exp Med Biol. 2018;1068: 33-43.
[10] WANG T, ZHENG X, LI R, et al. Integrated bioinformatic analysis reveals YWHAB as a novel diagnostic biomarker for idiopathic pulmonary arterial hypertension. J Cell Physiol. 2019;234(5):6449-6462.
[11] SU Y, TIAN X, GAO R, et al. Colon cancer diagnosis and staging classification based on machine learning and bioinformatics analysis. Comput Biol Med. 2022;145:105409.
[12] BARSHIR R, FISHILEVICH S, INY-STEIN T, et al. GeneCaRNA: a comprehensive gene-centric database of human non-coding RNAs in the GeneCards suite. J Mol Biol. 2021;433(11):166913.
[13] RITCHIE ME, PHIPSON B, WU D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43(7):e47.
[14] 徐文飞,明春玉,段戡,等.WGCNA和机器学习识别骨关节炎铁死亡特征基因及实验验证[J].中国组织工程研究,2024,28(30):4909-4914.
[15] LI Y, LU S, LAN M, et al. A prognostic nomogram integrating novel biomarkers identified by machine learning for cervical squamous cell carcinoma. J Transl Med. 2020;18(1):223.
[16] LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559.
[17] SUN Q, BAI L, ZHU S, et al. Analysis of lymphoma-related genes with gene ontology and kyoto encyclopedia of genes and genomes enrichment. Biomed Res Int. 2022;2022:8503511.
[18] HU X, NI S, ZHAO K, et al. Bioinformatics-led discovery of osteoarthritis biomarkers and inflammatory infiltrates. Front Immunol. 2022;13:871008.
[19] DHAWAN R, MOHANTY AK, KUMAR M, et al. Data from salivary gland proteome analysis of female Aedes aegypti Linn. Data Brief. 2017;13: 274-277.
[20] YU G, WANG LG, HAN Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284-287.
[21] LI Z, SILLANPÄÄ MJ. Overview of LASSO-related penalized regression methods for quantitative trait mapping and genomic selection. Theor Appl Genet. 2012;125(3):419-435.
[22] CHEN J, QI F, LI G, et al. Identification of the hub genes involved in stem cell treatment for intervertebral disc degeneration: a conjoint analysis of single-cell and machine learning. Stem Cells Int. 2023;2023:7055264.
[23] SANZ H, VALIM C, VEGAS E, et al. SVM-RFE: selection and visualization of the most relevant features through non-linear kernels. BMC Bioinformatics. 2018;19(1):432.
[24] HUANG X, ZENG J, ZHOU L, et al. A new strategy for analyzing time-series data using dynamic networks: identifying prospective biomarkers of hepatocellular carcinoma. Sci Rep. 2016;6:32448.
[25] ELLIS K, KERR J, GODBOLE S, et al. A random forest classifier for the prediction of energy expenditure and type of physical activity from wrist and hip accelerometers. Physiol Meas. 2014;35(11):2191-2203.
[26] GUO W, MU K, LI WS, et al. Identification of mitochondria-related key gene and association with immune cells infiltration in intervertebral disc degeneration. Front Genet. 2023;14:1135767.
[27] WU B, HUANG X, ZHANG M, et al. Significance of immune-related genes in the diagnosis and classification of intervertebral disc degeneration. J Immunol Res. 2022;2022:2616260.
[28] CHEN H, BOUTROS PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35.
[29] ROBIN X, TURCK N, HAINARD A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77.
[30] LIVAK KJ, SCHMITTGEN TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402-408.
[31] HÄNZELMANN S, CASTELO R, GUINNEY J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7.
[32] RUAN D, HE Q, DING Y, et al. Intervertebral disc transplantation in the treatment of degenerative spine disease: a preliminary study. Lancet. 2007;369(9566):993-999.
[33] YU L, HAO Y, PENG C, et al. Effect of Ginsenoside Rg1 on the intervertebral disc degeneration rats and the degenerative pulposus cells and its mechanism. Biomed Pharmacother. 2020;123:109738.
[34] XIN J, WANG Y, ZHENG Z, et al. Treatment of intervertebral disc degeneration. Orthop Surg. 2022;14(7):1271-1280.
[35] WANG J, XIA D, LIN Y, et al. Oxidative stress-induced circKIF18A downregulation impairs MCM7-mediated anti-senescence in intervertebral disc degeneration. Exp Mol Med. 2022;54(3):285-297.
[36] WEN P, ZHENG B, ZHANG B, et al. The role of ageing and oxidative stress in intervertebral disc degeneration. Front Mol Biosci. 2022;9:1052878.
[37] WANG Y, CHENG H, WANG T, et al. Oxidative stress in intervertebral disc degeneration:Molecular mechanisms, pathogenesis and treatment. Cell Prolif. 2023;56(9):e13448.
[38] VO N, NIEDERNHOFER LJ, NASTO LA, et al. An overview of underlying causes and animal models for the study of age-related degenerative disorders of the spine and synovial joints. J Orthop Res. 2013;31(6):831-837.
[39] CAO G, YANG S, CAO J, et al. The role of oxidative stress in intervertebral disc degeneration. Oxid Med Cell Longev. 2022;2022:2166817.
[40] VICTORELLI S, PASSOS JF. Reactive oxygen species detection in senescent cells. Methods Mol Biol. 2019;1896:21-29.
[41] TANG G, WANG Z, CHEN J, et al. Latent infection of low-virulence anaerobic bacteria in degenerated lumbar intervertebral discs. BMC Musculoskelet Disord. 2018;19(1):445.
[42] ASTUR N, MACIEL B, DOI AM, et al. Next-generation sequencing (NGS) to determine microbiome of herniated intervertebral disc. Spine J. 2022; 22(3):389-398.
[43] LI W, LAI K, CHOPRA N, et al. Gut-disc axis: a cause of intervertebral disc degeneration and low back pain? Eur Spine J. 2022;31(4):917-925.
[44] LI W, DING Z, ZHANG H, et al. The roles of blood lipid-metabolism genes in immune infiltration could promote the development of IDD. Front Cell Dev Biol. 2022;10:844395.
[45] SUN K, SUN X, SUN J, et al. Tissue renin-angiotensin system (tRAS) induce intervertebral disc degeneration by activating oxidative stress and inflammatory reaction. Oxid Med Cell Longev. 2021;2021:3225439.
[46] LI Z, WYSTRACH L, BERNSTEIN A, et al. The tissue-renin-angiotensin-system of the human intervertebral disc. Eur Cell Mater. 2020;40:115-132.
[47] GUO Y, GUO K, HU T, et al. Correlation between serum angiotensin-converting enzyme (ACE) levels and intervertebral disc degeneration. Peptides. 2022;157:170867.
[48] DEANE C, BROWN IR. Knockdown of heat shock proteins HSPA6 (Hsp70B’) and HSPA1A (Hsp70-1) sensitizes differentiated human neuronal cells to cellular stress. Neurochem Res. 2018;43(2):340-350.
[49] SHEN S, WEI C, FU J. RNA-sequencing reveals heat shock 70-kDa protein 6 (HSPA6) as a novel thymoquinone-upregulated gene that inhibits growth, migration, and invasion of triple-negative breast cancer cells. Front Oncol. 2021;11:667995.
[50] ZHANG HJ, LIAO HY, BAI DY, et al. MAPK /ERK signaling pathway: a potential target for the treatment of intervertebral disc degeneration. Biomed Pharmacother. 2021;143:112170.
[51] CAO S, LIU H, FAN J, et al. An oxidative stress-related gene pair (CCNB1/PKD1), competitive endogenous RNAs, and immune-infiltration patterns potentially regulate intervertebral disc degeneration development. Front Immunol. 2021;12:765382.
[52] QU Z, QUAN Z, ZHANG Q, et al. Comprehensive evaluation of differential lncRNA and gene expression in patients with intervertebral disc degeneration. Mol Med Rep. 2018;18(2):1504-1512.
[53] LIU D, WANG CJ, JUDGE DP, et al. A Pkd1-Fbn1 genetic interaction implicates TGF-β signaling in the pathogenesis of vascular complications in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2014;25(1):81-91.
[54] CHEN S, LEI L, LI Z, et al. Grem1 accelerates nucleus pulposus cell apoptosis and intervertebral disc degeneration by inhibiting TGF-β-mediated Smad2/3 phosphorylation. Exp Mol Med. 2022;54(4):518-530.
[55] CHEN S, LIU S, MA K, et al. TGF-β signaling in intervertebral disc health and disease. Osteoarthritis Cartilage. 2019, 27(8):1109-1117.
[56] WU T, LI X, JIA X, et al. Krüppel like factor 10 prevents intervertebral disc degeneration via TGF-β signaling pathway both in vitro and in vivo. J Orthop Translat. 2021;29:19-29.
[57] RISBUD MV, SHAPIRO IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10(1):44-56.
[58] LI J, YU C, NI S, et al. Identification of core genes and screening of potential targets in intervertebral disc degeneration using integrated bioinformatics analysis. Front Genet. 2022;13:864100.
[59] LAN T, HU Z, GUO W, et al. Development of a novel inflammatory-associated gene signature and immune infiltration patterns in intervertebral disc degeneration. Oxid Med Cell Longev. 2022;2022:2481071.
[60] PENG B, HAO J, HOU S, et al. Possible pathogenesis of painful intervertebral disc degeneration. Spine (Phila Pa 1976). 2006;31(5):560-566.
[61] KEPLER CK, MARKOVA DZ, DIBRA F, et al. Expression and relationship of proinflammatory chemokine RANTES/CCL5 and cytokine IL-1β in painful human intervertebral discs. Spine (Phila Pa 1976). 2013;38(11):873-880. |