中国组织工程研究 ›› 2021, Vol. 25 ›› Issue (35): 5716-5722.doi: 10.12307/2021.305
• 组织构建综述 tissue construction review • 上一篇 下一篇
表观遗传学对牙周疾病及牙周细胞成骨和骨再生的作用机制
吴丹丹,刘 琪
- 遵义医科大学附属口腔医院,贵州省遵义市 563000
-
收稿日期:2021-01-04修回日期:2021-01-06接受日期:2021-02-18出版日期:2021-12-18发布日期:2021-08-05 -
通讯作者:刘琪,博士,教授,遵义医科大学附属口腔医院,贵州省遵义市 563000 -
作者简介:吴丹丹,女,1995年生,贵州省绥阳县人,汉族,硕士,主要从事2型糖尿病与牙周炎之间关系的研究 -
基金资助:国家自然科学基金项目(8186040218),项目负责人:刘琪;国家自然科学基金项目(81760199);贵州省科技计划项目(黔科合基础[2020]1Y328,黔科合基础[2018]1185)
Epigenetic mechanisms in periodontal disease, periodontal cell osteogenesis and bone regeneration
Wu Dandan, Liu Qi
- Stomatological Hospital Affiliated to Zunyi Medical University, Zunyi 563000, Guizhou Province, China
-
Received:2021-01-04Revised:2021-01-06Accepted:2021-02-18Online:2021-12-18Published:2021-08-05 -
Contact:Liu Qi, MD, Professor, Stomatological Hospital Affiliated to Zunyi Medical University, Zunyi 563000, Guizhou Province, China -
About author:Wu Dandan, Master, Stomatological Hospital Affiliated to Zunyi Medical University, Zunyi 563000, Guizhou Province, China -
Supported by:the National Natural Science Foundation of China, No. 8186040218 (to LQ) and 81760199; Guizhou Provincial Science and Technology Plan Projects, No. [2020]1Y328 and [2018]1185
摘要:
文题释义:
表观遗传学:是研究基因的核苷酸序列不发生改变的情况下,基因表达的可遗传变化的一门遗传学分支学科。表观遗传的现象很多,已知的有DNA甲基化(DNA methylation)、基因组印记(genomic imprinting)、母体效应(maternal effects)、基因沉默(gene silencing)、核仁显性、休眠转座子激活和RNA编辑(RNA editing)等。
DNA甲基化(DNA methylation):为DNA化学修饰的一种形式,能够在不改变DNA序列的前提下改变遗传表现。所谓DNA甲基化是指在DNA甲基化转移酶的作用下,在基因组CpG二核苷酸的胞嘧啶5’碳位共价键结合一个甲基基团。大量研究表明,DNA甲基化能引起染色质结构、DNA构象、DNA稳定性及DNA与蛋白质相互作用方式的改变,从而控制基因表达。
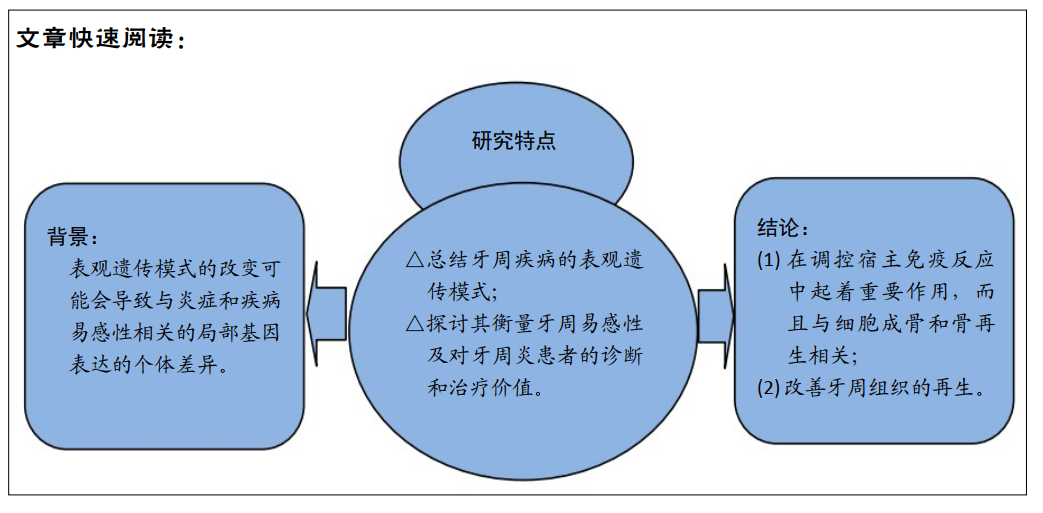
背景:牙周疾病主要是由细菌引起的复杂的、异质的和多因素的口腔破坏性疾病,而表观遗传在其中发挥的作用不可忽视。
目的:总结牙周疾病的表观遗传模式,探讨其衡量牙周易感性及对牙周炎患者的诊断和治疗价值。
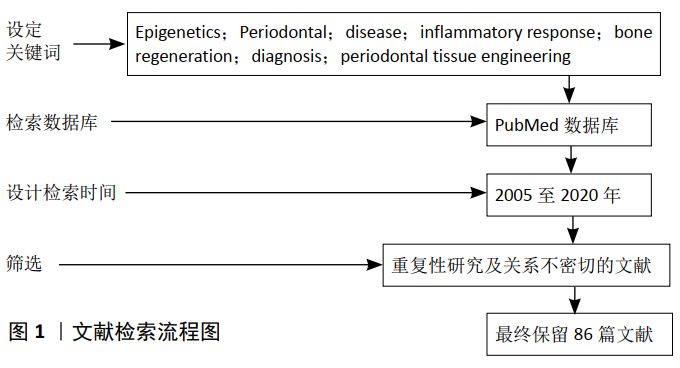
方法:以PubMed数据库为主检索2005至2020年的有关文献;检索词为:Epigenetics;Periodontal disease;inflammatory response;bone regeneration;diagnosis;periodontal tissue engineering。经过筛选,最终对86篇相关文献进行了探讨。
结果与结论:牙周疾病与表观遗传两者之间相互影响,口腔病原体对病原识别受体(例如Toll样受体)以及促炎/抗炎细胞因子的表观遗传学调控影响的发现表明,其在调控宿主免疫反应中起着重要作用,而且与细胞成骨和骨再生相关。组织工程学方面的最新研究表明表观遗传学也可以改善牙周组织的再生。
https://orcid.org/0000-0002-3556-9219 (吴丹丹)
中图分类号:
引用本文
吴丹丹, 刘 琪. 表观遗传学对牙周疾病及牙周细胞成骨和骨再生的作用机制[J]. 中国组织工程研究, 2021, 25(35): 5716-5722.
Wu Dandan, Liu Qi. Epigenetic mechanisms in periodontal disease, periodontal cell osteogenesis and bone regeneration[J]. Chinese Journal of Tissue Engineering Research, 2021, 25(35): 5716-5722.
2.1.1 DNA甲基化 DNA甲基化是主要的表观遗传修饰,其中二核苷酸胞嘧啶-磷酸-鸟嘌呤(CpG)中的胞嘧啶(C)被甲基(–CH3)共价修饰,导致“第五DNA序列中的5’甲基胞嘧啶(5mC)为碱基。DNA甲基转移酶(DNA methyltransferase,DNMT)可催化这种修饰。DNMT1负责维持现有的DNA甲基化,并且从头甲基转移酶DNMT3A和DNMT3B作用于半甲基化和/或未甲基化的CpG位点,建立新的甲基化模式[9]。
2.1.2 组蛋白修饰 组蛋白是核小体的核心成分,负责稳定地维持抑制性染色质。核小体由DNA链组成,该DNA链缠绕在八聚体核心中的两个组蛋白H2A,H2B,H3和H4的两个副本中,每个核心带有一个连接子组蛋白H1,染色质由核小体的重复亚基组成,并有可能定义细胞内遗传信息的结构状态。染色质结构的构象变化赋予基因组特定的排列,处于浓缩或非浓缩状态,从而改变和控制基因表达。组蛋白修饰是指组蛋白甲基化、乙酰化、磷酸化、聚腺苷酸化、泛素化和ADP核糖基化的过程[10]。这些修饰与多种酶如:组蛋白乙酰转移酶(HAT)和组蛋白去乙酰基酶(histone deacetylase,HDAC) 以及可以修饰组蛋白尾部氨基酸残基的组蛋白甲基转移酶(HMT)和脱甲基酶(HDM)有关[5]。组蛋白乙酰化与转录激活有关[11],而组蛋白甲基化则与差异基因表达相关(即活性转录或阻抑),这取决于组蛋白尾巴上甲基赖氨酸残基的位置和甲基化程度。
2.1.3 ncRNA ncRNA参与控制基因表达、染色质结构调节、转录调控和转录后修饰[12]。ncRNA按长度分类,基本上分为微小RNA(miRNA,19-31 nt)和长ncRNA(lncRNA,> 200 nt)[13]。它们通过与靶mRNA的3’非翻译区结合而调节基因表达从而发挥抑制作用,这可能是由于目标mRNA的降解或通过阻止翻译引起的[14]。新发现的甲基化作用是调节信使RNA(mRNA),最重要和已知的转录组修饰是N6-甲基腺苷(m6A)[15]。
2.2 牙周疾病与表观遗传 牙周炎是一种感染性疾病,其特征是细菌定植在牙齿表面后,牙龈组织出现慢性炎症,最终导致组织破坏、牙槽骨和结缔组织丢失。而口腔中各种病原体的暴露、信号的激活、促炎与抗炎分子的聚集,可诱导炎症性反应。表观遗传改变发生在牙齿周围的生物膜-牙龈界面处,并且在同一个体的牙周发炎部位和非发炎部位之间,表观遗传修饰有所不同[3,16]。感染和宿主的免疫反应可能会导致表观基因组发生变化,进而可能增加对疾病的敏感性。免疫应答中,如细菌因素作为外部刺激启动细胞内级联信号,激活转录结合DNA调控序列的因素,从而诱导某些基因的表达[17-18]。
2.2.1 口腔病原体入侵引发牙周细胞表观遗传改变 口腔上皮细胞是抵抗病原体的一线防御,可以假设细菌的存在会诱导这些细胞中表观基因组的改变,进而通过诱导信号传导途径和基因表达的变化而影响炎症细胞。牙龈卟啉单胞菌(P.gingivalis)和核梭状芽胞杆菌(F.nucleatum)能够诱导组蛋白乙酰化并下调DNMT1,密螺旋体(T.denticola)使得牙周膜细胞中几个Ⅱ类HDAC家族成员,包括HDAC4,HDAC6和HDAC10的转录水平下调。YIN等[19]研究通过表观遗传调控提供了细菌特异性先天免疫应答的见解,细菌的存在会导致牙龈上皮的表观遗传修饰。牙龈卟啉单胞菌刺激导致DNMT1和HDAC1和HDAC2的水平降低,致使牙龈上皮细胞中DNA和组蛋白发生化学变化。此外,这些细菌对病原体识别受体(PRRs)和Toll样受体(TLRs)的激活进一步诱导了口腔上皮细胞中的组蛋白修饰[10,20]。Toll样受体通过识别包括细菌脂多糖在内的所谓病原体相关分子模式的能力,在先天免疫中发挥重要作用。Toll样受体表达的失调可能影响宿主对牙周病原体的反应,导致炎症增加和对牙周炎的易感性[21],在人牙龈上皮细胞中研究了Toll样受体2的DNA甲基化模式表明牙龈卟啉单胞菌介导了牙龈的DNA甲基化,在用牙龈卟啉单胞菌处理的小鼠牙龈中也有类似的发现。据报道,miRNA可能通过调节丝裂原活化蛋白激酶(MAPK)介导内毒素耐受性,当暴露于细菌脂多糖或其靶标时增加Toll样受体的敏感性[22]。另外,细胞表面受体表达的降低影响了细胞对细胞外基质的黏附,进而导致基质金属蛋白酶表达的开始,当牙龈卟啉单胞菌衍生的脂多糖刺激人牙周膜成纤维细胞时,观察到25个细胞外基质相关基因在启动子区域甲基化。在实验中还发现脂多糖可能抑制细胞外基质的重塑,从而通过DNA甲基化改变影响成骨分化[23]。基质金属蛋白酶是基质降解和骨吸收以及伤口愈合、细胞增殖、炎症和免疫力的关键因素,有发现表明密螺旋体可诱导基质金属蛋白酶2启动子的甲基化不足以及牙周膜细胞中前基质金属蛋白酶2的慢性活化,这表明密螺旋体通过表观遗传机制在牙周炎中支持组织的丧失中发挥了作用[24-25]。口腔病原体还分泌丁酸、异丁酸等物质抑制HDAC的表达、促进组蛋白乙酰化,而这会影响中性粒细胞的功能,促进伴放线聚集杆菌的感染[26]。高浓度丁酸还可通过抑制HDAC来破坏骨保护素/核因子κB受体活化因子配体的平衡,参与牙周及根尖周组织的骨质破坏过程[27]。
2.2.2 牙周炎中细胞因子的表观遗传变化 牙周组织中的炎症反应诱导表观遗传修饰并改变细胞因子基因表达;DNA高甲基化和组蛋白去乙酰化可抑制细胞因子的表达,而DNA低甲基化和组蛋白乙酰化则能促进基因的表达;持续性炎症导致DNA甲基化和细胞因子信号转导抑制剂的沉默,同时它诱导细胞因子信号转导的激活[28-29];促炎和抗炎细胞因子、趋化因子作用于发炎的牙周组织,例如白细胞介素6、白细胞介素6R、白细胞介素1β、白细胞介素10、趋化因子配体8、肿瘤坏死因子和细胞因子信号抑制物1(suppressors of cytokine signaling 1,SOCS1)等炎症候选基因的作用已在牙周病患者中得到证实。
(1)白细胞介素6:是一种多功能的促炎细胞因子,在牙周炎患者中高表达。KOBAYASHI等[30]分析慢性牙周炎组与健康组牙龈组织和外周血中白细胞介素6的DNA甲基化谱和白细胞介素6的mRNA表达,与外周血相比,慢性牙周炎组牙龈组织中包含19个CpG基序的白细胞介素6启动子的总体甲基化率显著降低,慢性牙周炎患者牙龈组织中白细胞介素6基因转录表达的增加可能与白细胞介素6启动子的低甲基化有关。
(2)白细胞介素6R:SCHULZ等[31]报道侵袭性牙周炎组和对照组之间白细胞介素6R甲基化没有统计学上的显著差异。而相比之下,在局限性侵袭性牙周炎研究中,SHADDOX等[32]观察到与健康组相比,中度局限性侵袭性牙周炎和严重局限性侵袭性牙周炎中白细胞介素6R甲基化的统计学差异显著增加。
(3)白细胞介素1β:SEUTTER等[33]研究显示成纤维细胞暴露于白细胞介素1β和前列腺素E2会导致DNA甲基化/去甲基化酶表达改变以及DNA甲基化模式改变。
(4)白细胞介素10:是一种抗炎细胞因子,由淋巴细胞、单核细胞和巨噬细胞分泌。尽管在慢性牙周炎和健康个体之间未观察到白细胞介素10甲基化的差异[34],这表明白细胞介素10启动子的超甲基化是健康和疾病的共同特征,但作者仍然报告白细胞介素10基因中存在部分甲基化,这一发现可能是由于在激活的不同阶段存在各种炎症细胞而引起的。白细胞介素10基因多态性与牙周炎有关[35],在最近的荟萃分析中也表明白细胞介素10多态性与慢性牙周炎的易感性有关[36]。
(5)趋化因子配体8:是一种重要的趋化因子[37],在牙周疾病中起着重要作用,因为它募集并激活了急性炎症细胞。从表观遗传学的角度来看,侵袭性牙周炎患者的趋化因子配体8明显低甲基化,提示趋化因子配体8是侵袭性牙周炎发病机制中的潜在病因[38]。SCHULZ等[31]分析侵袭性牙周炎患者和健康受试者牙龈组织中22种炎性基因的DNA甲基化水平,并观察到牙周炎患者中白细胞介素17C和CCL25的甲基化程度较低。CpG甲基化的减少可能伴随着基因表达的增加,这可能导致白细胞介素17C、CCL25的可用性更高,随后牙周附着丧失。
(6)肿瘤坏死因子α:是在活动性和进行性牙周炎中升高的原发性炎性细胞因子。有研究报道了慢性牙周炎中肿瘤坏死因子甲基化增加[39],这导致该疾病中肿瘤坏死因子α的表达降低。因此,慢性牙周炎中肿瘤坏死因子的高甲基化可能是调节性的抑制机制,可以保护宿主免受长时间的环境刺激。相反,LAVU等[40]报道牙周炎组的肿瘤坏死因子启动子甲基化水平低于对照组,这些研究之间发现的差异可能由于检查的甲基化区域的不同。最近,发现miRNA-142被肿瘤坏死因子α上调,并可以通过抑制BACH2表达来介导肿瘤坏死因子α诱导牙周炎的牙龈上皮细胞凋亡[41]。
(7)细胞因子信号抑制物1:它是最强的细胞因子信号传导抑制剂之一[42]。与侵袭性牙周炎相比,细胞因子信号抑制物1在健康受试者中表现出更多的低甲基化[43]。细胞因子信号抑制物1的发现很有趣,因为它是控制炎症的基因,因此,其甲基化模式可能参与了侵袭性牙周炎的发展。
2.3 牙周细胞调节骨骼再生的表观遗传 来自牙周膜的细胞具有分化为成骨细胞的潜力,而 Runt相关转录因子 (runt-related transcription factor 2,RUNX2)是该过程的关键因素。牙龈卟啉单胞菌脂多糖诱导人牙周膜细胞中DNMT1的增加和RUNX2表达的下调。这表明,观察到的脂多糖对成骨细胞分化的抑制作用可能是RUNX2的DNA甲基化不足的结果[44]。在CHO等[45]通过诱导成骨因子RUNX2和碱性磷酸酶的去甲基化及基因表达,诱导人牙龈成纤维细胞分化为成骨细胞[46]。用DNA甲基化抑制剂诱导的RUNX2和碱性磷酸酶的甲基化不足来治疗人牙龈成纤维细胞,然后用骨形态发生蛋白2处理这些细胞,导致成纤维细胞向成骨细胞谱系和骨形成分化。HUYNH等[46]证明,组蛋白去乙酰化酶抑制剂(histone deacetylase inhibitor,HDACi)曲古抑菌素A(TSA)处理人牙周膜细胞,可诱导成骨分化并增加成骨细胞相关基因的表达。此外,曲古抑菌素A诱导的组蛋白H3的超乙酰化会降低成骨分化过程中HDAC3的表达,并促进人牙周膜细胞中RUNX2的表达。这些结果表明,这种方法可能代表了一种新的牙槽骨再生治疗方法。体细胞重编程技术也在其中发挥作用;有研究通过表观遗传修饰从非成骨细胞到成骨细胞的直接转分化研究骨再生,表观遗传修饰可以允许将非成骨细胞直接编程为成骨细胞,这种方法可能是骨再生的新治疗途径[47]。
2.4 表观遗传学在牙周领域的临床应用
2.4.1 表观遗传学作为牙周炎的潜在诊断 表观遗传学有助于更好地理解基因与环境之间的相互作用,并可以解释为什么具有相同临床表型的患者而治疗反应却不同[48]。DNA甲基化状态的改变与炎症有关,目前被评估为多种疾病的潜在生物标记物,包括克罗恩病和狼疮[49-50]。巨噬细胞是免疫系统的关键免疫细胞,在维持牙周动态平衡方面起着至关重要的调节作用。JIANG等[51]发现DNA甲基化改变可能导致与牙周疾病有关的基因表达异常,从而导致巨噬细胞失控,这为将DNA甲基化相关参数作为牙周炎的诊断和治疗新的生物标志物提供了前景。在牙周炎患者中,启动子特异性分析和EWAS 均已鉴定出血液样本中DNA甲基化标记改 变[52-53],这可用于诊断目的,但是它们与疾病易感性、进展或严重性之间的关系尚待评估,这对于考虑将其用作疾病生物标记物之前是很有必要的。其次,能将表观遗传模式/标记物与临床表型相联系起来,并且将表观遗传学作为鉴定患有牙周炎风险的工具,比如颊拭子刮擦口腔黏膜或唾液、龈沟液进行表观遗传分析,使其作为临床上的诊断工具,与周围血液样本相比,颊组织与病理口腔微生物群之间的相互作用更为密切[54],它可以提供有关疾病发病机制的更多相关信息;唾液则是作为“液体活检”,DNA甲基化也已经在口腔拭子和唾液中得以研究[55-56]。目前,也有表观遗传学分析与临床测量相结合的研究。牙周炎患者牙龈组织中白细胞介素6的甲基化水平与探诊深度之间呈负相关[51],而与Toll样受体2甲基化和探诊深度呈正相关[57]。2019年的一项研究表明肿瘤坏死因子α与牙周临床指标探诊出血呈显著正相关,表明肿瘤坏死因子αDNA甲基化可能是牙周健康的生物标志物[58]。
2.4.2 表观遗传药物作为牙周疾病潜在治疗靶标 表观遗传修饰显然与牙周疾病的发展有关,并且目前针对与组织稳态破坏和牙周炎发展相关的表观遗传靶标的新兴治疗[59]。表观遗传的可逆性也使之成为癌症和炎症性疾病新治疗模型的有吸引力的方向。“表观药物”一词是Ivanov及其同事创造的,它们是抑制或激活与疾病相关的表观遗传蛋白,可改善、治愈或预防疾病的药物[60]。在癌症领域中,有许多关于使用表观药物的研究,并且最近有营养作为治疗模型,但是目前缺乏关于与口腔健康有关的表观药物的研究。在有关HDACi对骨重塑影响的综述中报道其影响破骨细胞的分化、成熟和活性[61]。牙周组织再生的挑战在于它既涉及细胞分化为成骨细胞,又涉及减少炎症。在实验性牙周炎中,用溴结构域和末端外结构域(bromodomain and extra-terminal domain,BET)抑制剂JQ1处理小鼠会降低牙槽骨吸收和炎性细胞因子的表达[62]。有人建议将表观药物作为改善组织再生的工具,用HDACi丁酸钠治疗牙周膜成纤维细胞可促进成骨细胞相关蛋白的表达,并抑制促炎细胞因子的产生[63]。已经证明HDACi 1179.4b可以抑制牙槽骨丢失,但不能抑制牙龈发炎[64]。相反,使用BET抑制剂JQ1可以抑制炎症反应和牙槽骨丢失。BET溴结构域蛋白是重要的基因表达表观遗传调节剂,可与乙酰化组蛋白相互作用,从而影响转录机制从而调节基因转录[62]。牙龈成纤维细胞和牙龈上皮细胞系端粒酶永生化牙龈角质形成细胞中的BET抑制剂I-BET151和JQ1抑制了炎症基因的表达阻止牙龈组织中的驻留细胞过度产生炎症递质,将BET蛋白家族确定为牙周疾病的潜在治疗靶标[65]。
在炎症细胞中发现了HDAC1蛋白,提示其在炎症调节中的作用。使用DNA甲基化抑制剂5-Aza-2’脱氧胞苷可以提高牙龈成纤维细胞对转化生长因子β1的反应性,并提高DNMT的水平,这表明它是改善伤口愈合和牙周组织再生的新潜在工具[66]。基于单药治疗的特征在于特定靶标的参与而具有选择性作用机制的特点,尤其是在多因素疾病中,这还远远不够;具体而言,在不同的复杂疾病(例如癌症,神经退行性疾病和代谢性疾病)中,能够同时击中两个或多个靶标的杂交药物的开发,其中一个(或多个)是表观遗传的而另一个如双重组蛋白脱乙酰基酶/激酶抑制剂所示,属于非成因类别的结果非常有前途,两种化合物CUDC-101和CUDC-907已进入临床试验[67]。但是,针对牙周疾病患者的表观遗传修饰的尝试需要谨慎进行,癌症患者的临床试验证据表明,广谱HDACi具有多种剂量限制性毒性,包括血小板减少症、中性粒细胞减少症、疲劳和腹泻[68]。关于BET抑制剂在牙周炎中的潜在应用,需要解决类似的问题。这些化合物已进入癌症临床试验[69],但尚未在具有任何炎性疾病的患者中进行评估,其对宿主与病原体相互作用的影响尚待确定。
2.5 牙周组织工程 牙周炎中支持组织的丧失是一项重大的临床挑战。为了改善牙周炎预后,需要牙槽骨的再生。牙周组织工程的目标是恢复所有牙周成分,即牙槽骨、牙周韧带、牙骨质和周围的软组织[70],而用于组织再生的两个主要工具是种子细胞和生物支架材料。
2.5.1 种子细胞与表观遗传 种子细胞是进行牙周组织再生治疗的关键,而β-Catenin、骨形态发生蛋白2和RUNX2是干细胞分化调控的关键蛋白,它们的表达受到表观遗传的干扰[71];Runx2是牙源性间充质干细胞分化中的主要转录因子,其表达受不同表观遗传因子控制[72-73]。脂多糖对RUNX2表达的这种抑制作用与H3K27me3对基因启动子的富集增加有关,在其他牙源性祖细胞(包括牙囊和牙髓干细胞),在基质和矿化基因处也观察到类似的组蛋白甲基化模式[74]。基因表达的牙源性祖细胞特异性激活途径可能使这些细胞根据环境线索在牙齿发育和再生过程中产生未矿化或矿化的牙齿组织;与之相印证的是在高糖条件下,体外应用5-Aza-2’脱氧胞苷降低人牙周膜干细胞中的DNA甲基化水平能够挽救这些细胞的成骨能力[75]。
报告显示,曲古抑菌素A促进脂肪干细胞的成骨分化,增强组蛋白H3乙酰化,并增强RUNX2的表达[76],并且研究表明赖氨酸特异性脱甲基酶1(lysine-specific demethylase 1,LSD1,一种组蛋白脱甲基酶),可特异性催化二甲基和单甲基组蛋白H3赖氨酸4(H3K4me2/1)的脱甲基化,作为人脂肪来源的干细胞成骨分化的关键调控因子,LSD1脱甲基酶抑制剂可以有效地阻断其催化活性,并在表观遗传上促进人脂肪来源的干细胞的成骨分化[77],对LSD1如何影响成骨相关表观遗传事件的更好理解将为基于人脂肪来源的干细胞疗法的调节提供新见解,并通过表观遗传干预改善骨组织工程的发展。从牙周的观点来看,增强脂肪干细胞的成骨分化非常重要,因为这些干细胞已经显示出牙周组织再生的潜力。此外,长链非编码 RNA(long non-coding RNA,LncRNA)人母系表达基因(MEG3)或果蝇 zeste 基因增强子的人类同源物 2(enhancer of zeste homolog 2,EZH2,组蛋白甲基转移酶之一)的减少通过表观遗传调控Wnt基因启动子上的H3K27me3水平激活了Wnt/β-catenin信号通路[78]。此外还有研究表明lncRNA 核仁小分子 RNA 宿主基因1(small nucleolar RNA host gene 1,SNHG1)通过EZH2介导的Kruppel样因子2 (kruppel-like factor 2,KLF2)启动子的H3K27me3甲基化抑制牙周膜干细胞的成骨分化,表明SNHG1-KLF2轴具有作为骨再生的新型治疗靶标的巨大潜力[79]。这些研究为牙周组织工程和成骨组织再生提供了新的目标。
2.5.2 生物支架材料的表观遗传 支架的功能是提供附着平台,以指导组织并阻止上皮细胞的向下生长,支架的结构、外形和材料特性会影响这些过程;此外,这些因素还影响细胞向支架的分化、迁移和黏附[80],这是确保组织再生至关重要的因素。最近,关于材料以及表面结构对表观遗传机制影响的研究正在涌现出来。在坚硬材料表面上生长的细胞具有转录活性染色质,而在软质材料上生长的细胞具有转录惰性染色质[81]。因此,在组织工程中引入表观遗传学作为新的治疗选择。研究表明,在纳米级层面改变二氧化钛的表面结构会改变人脂肪细胞中的组蛋白甲基化模式,从而将这些细胞引导向成骨细胞分化[82]。
人工合成的羟基磷灰石被认为是一种适用于支架和植入物涂层的潜在生物材料。研究表明,暴露于纳米级羟基磷灰石颗粒的骨髓基质细胞和成骨前细胞的DNA甲基化模式和碱性磷酸酶的基因表达发生了变化;分化早期的成骨细胞比后期的成骨细胞更易受感染[83]。有人建议采用纳米结构形貌的支架作为改善牙周组织工程的潜在工具[84]。尽管关于表面形貌和物质能量如何影响表观基因组的研究尚处于起步阶段,但当前的研究表明,使用材料和纳米技术通过表观遗传学促进组织再生和细胞功能的可能性很大。它还显示了具有细胞功能和物质结构的重要性,以便获得最佳的牙周再生结果。最近有研究小组使用含锶的生物活性玻璃作为支架,发现剪接因子异质核糖核糖核蛋白L(hnRNPL)在锶(Sr)通过激活AKT途径刺激的牙周膜干细胞中被大幅下调。此外,异质核糖核糖核蛋白L通过下调H3K36me3特异性甲基转移酶Setd2来抑制牙周膜干细胞的成骨分化[85]。支架的结构不仅可以诱导表观遗传学改变,还可以将支架用作表观药物的递送模型。二氧化硅是一种已被FDA批准为DNA甲基化抑制剂5’-氮杂的载体[86]。通过将支架特性与表观药物的递送结合起来,可以进一步改善组织再生。此外,在体内小鼠模型中,这些细胞与Bio-Oss骨材料和Tisseel纤维蛋白凝胶一起移植,与对照相比,导致骨矿物质含量和骨形成增加。

| [1] BORRELL LN, PAPAPANOU PN. Analytical epidemiology of periodontitis. J Clin Periodontol. 2005;32(Suppl 6):132-158. [2] BLOCK T, EL-OSTA A. Epigenetic programming, early life nutrition and the risk of metabolic disease. Atherosclerosis. 2017;266(12):31-40. [3] LARSSON L, CASTILHO RM, GIANNOBILE WV. Epigenetics and its role in periodontal diseases: a state-of-the-art review. J Periodontol. 2015;86(4):556-568. [4] WADDINGTON CH. The epigenotype. 1942. Int J Epidemiol. 2012;41(1): 10-13. [5] MIRANDA FURTADO CL, DOS SANTOS LUCIANO MC, SILVA SANTOS RD, et al. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics. 2019;14(12):1164-1176. [6] BAYARSAIHAN D. Epigenetic mechanisms involved in modulation of inflammatory diseases. Curr Opin Clin Nutr Metab Care. 2016;19(4): 263-269. [7] HERCEG Z. Epigenetic Mechanisms as an Interface Between the Environment and Genome. Adv Exp Med Biol. 2016;903(1):3-15. [8] PERKINS DJ, PATEL MC, BLANCO JC, et al. Epigenetic Mechanisms Governing Innate Inflammatory Responses. J Interferon Cytokine Res. 2016;36(7):454-461. [9] LYKO F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19(2):81-92. [10] MARTINS MD, JIAO Y, LARSSON L, et al. Epigenetic Modifications of Histones in Periodontal Disease. J Dent Res. 2016;95(2):215-222. [11] JAVAID N,CHOI S. Acetylation- and Methylation-Related Epigenetic Proteins in the Context of Their Targets. Genes. 2017;8(8):196-196. [12] PENNA I, GIGONI A, COSTA D, et al. The inhibition of 45A ncRNA expression reduces tumor formation, affecting tumor nodules compactness and metastatic potential in neuroblastoma cells. Oncotarget. 2017;8(5):8189-8205. [13] QI P, ZHOU XY, DU X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15(1):39-39. [14] FILIPOWICZ W, BHATTACHARYYA SN, SONENBERG N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?. Nat Rev Genet. 2008;9(2):102-114. [15] VISVANATHAN A, SOMASUNDARAM K. mRNA Traffic Control Reviewed: N6-Methyladenosine (m(6) A) Takes the Driver’s Seat. Bioessays. 2018; 40(1):1-12. [16] BARROS SP, OFFENBACHER S. Modifiable risk factors in periodontal disease: epigenetic regulation of gene expression in the inflammatory response. Periodontol 2000. 2014;64(1):95-110. [17] RINN JL, CHANG HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145-166. [18] SMALE ST. Transcriptional regulation in the innate immune system. Curr Opin Immunol. 2012;24(1):51-57. [19] YIN L, CHUNG WO. Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol. 2011;4(4):409-419. [20] ATEIA IM, SUTTHIBOONYAPAN P, KAMARAJAN P, et al. Treponema denticola increases MMP-2 expression and activation in the periodontium via reversible DNA and histone modifications. Cell Microbiol. 2018; 20(4):1-60. [21] BENAKANAKERE M, ABDOLHOSSEINI M, HOSUR K, et al. TLR2 promoter hypermethylation creates innate immune dysbiosis. J Dent Res. 2015; 94(1):183-191. [22] NA HS, PARK MH, SONG YR, et al. Elevated MicroRNA-128 in Periodontitis Mitigates Tumor Necrosis Factor-α Response via p38 Signaling Pathway in Macrophages. J Periodontol. 2016;87(9):e173-e182. [23] TAKAI R, UEHARA O, HARADA F, et al. DNA hypermethylation of extracellular matrix-related genes in human periodontal fibroblasts induced by stimulation for a prolonged period with lipopolysaccharide derived from Porphyromonas gingivalis. J Periodontal Res. 2016;51(4):508-517. [24] MIAO D, GODOVIKOVA V, QIAN X, et al. Treponema denticola upregulates MMP-2 activation in periodontal ligament cells: interplay between epigenetics and periodontal infection. Arch Oral Biol. 2014; 59(10):1056-1064. [25] FRANCO C, PATRICIA HR, TIMO S, et al. Matrix metalloproteinases as regulators of periodontal inflammation. Int J Mol Sci. 2017;18(2):440-440. [26] CORRÊA RO, VIEIRA A, SERNAGLIA E, et al. Bacterial short‐chain fatty acid metabolites modulate the inflammatory response against infectious bacteria. Cellular microbiology. 2017;19(7):1-47. [27] CHANG MC, CHEN YJ, LIAN YC, et al. Butyrate Stimulates Histone H3 Acetylation, 8-Isoprostane Production, RANKL Expression, and Regulated Osteoprotegerin Expression/Secretion in MG-63 Osteoblastic Cells. Int J Mol Sci. 2018;19(12):4071-4071. [28] SEO JY, PARK YJ, YI YA, et al. Epigenetics: general characteristics and implications for oral health. Restor Dent Endod. 2015;40(1):14-22. [29] LUO Y, PENG X, DUAN D, et al. Epigenetic Regulations in the Pathogenesis of Periodontitis. Curr Stem Cell Res Ther. 2018;13(2):144-150. [30] KOBAYASHI T, ISHIDA K,YOSHIE H. Increased expression of interleukin-6 (IL-6) gene transcript in relation to IL-6 promoter hypomethylation in gingival tissue from patients with chronic periodontitis. Arch Oral Biol. 2016;69(3):89-94. [31] SCHULZ S, IMMEL UD, JUST L, et al. Epigenetic characteristics in inflammatory candidate genes in aggressive periodontitis. Hum Immunol. 2016;77(1):71-75. [32] SHADDOX LM, MULLERSMAN AF, HUANG H, et al. Epigenetic regulation of inflammation in localized aggressive periodontitis. Clin Epigenetics. 2017;9(23):94-94. [33] SEUTTER S, WINFIELD J, ESBITT A, et al. Interleukin 1β and Prostaglandin E2 affect expression of DNA methylating and demethylating enzymes in human gingival fibroblasts. Int Immunopharmacol. 2020;78(11):1-10. [34] VIANA MB, CARDOSO FP, DINIZ MG, et al. Methylation pattern of IFN-γ and IL-10 genes in periodontal tissues. Immunobiology. 2011;216(8): 936-941. [35] ZHANG Q, CHEN B, YAN F, et al. Interleukin-10 inhibits bone resorption: a potential therapeutic strategy in periodontitis and other bone loss diseases. Biomed Res Int. 2014;2014(10):1-5. [36] ZHANG Z, ZHENG Y, LI X. Interleukin-10 gene polymorphisms and chronic periodontitis susceptibility: Evidence based on 33 studies. J Periodontal Res. 2019;54(2):95-105. [37] FINOTI LS, NEPOMUCENO R, PIGOSSI SC, et al. Association between interleukin-8 levels and chronic periodontal disease: A PRISMA-compliant systematic review and meta-analysis. Medicine (Baltimore). 2017;96(22):1-8. [38] ANDIA DC, DE OLIVEIRA NF, CASARIN RC, et al. DNA methylation status of the IL8 gene promoter in aggressive periodontitis. J Periodontol. 2010;81(9):1336-1341. [39] ZHANG S, BARROS SP, MORETTI AJ, et al. Epigenetic regulation of TNFA expression in periodontal disease. J Periodontol. 2013;84(11):1606-1616. [40] LAVU V, VETTRISELVI V, PRIYANKA V, et al. Methylation Status of Promoter Region of Tumor Necrosis Factor Alpha Gene in Subjects with Healthy Gingiva and Chronic Periodontitis–A Pilot Study. Biomed Pharmacol J. 2019;12(2):639-647. [41] LI S, SONG Z, DONG J, et al. microRNA-142 is upregulated by tumor necrosis factor-alpha and triggers apoptosis in human gingival epithelial cells by repressing BACH2 expression. Am J Transl Res. 2017;9(1):175-183. [42] BABON JJ, NICOLA NA. The biology and mechanism of action of suppressor of cytokine signaling 3. Growth Factors. 2012;30(4):207-219. [43] BAPTISTA NB, PORTINHO D, CASARIN RC, et al. DNA methylation levels of SOCS1 and LINE-1 in oral epithelial cells from aggressive periodontitis patients. Arch Oral Biol. 2014;59(7):670-678. [44] UEHARA O, ABIKO Y, SAITOH M, et al. Lipopolysaccharide extracted from Porphyromonas gingivalis induces DNA hypermethylation of runt-related transcription factor 2 in human periodontal fibroblasts. J Microbiol Immunol Infect. 2014;47(3):176-181. [45] CHO Y, KIM B, BAE H, et al. Direct Gingival Fibroblast/Osteoblast Transdifferentiation via Epigenetics. J Dent Res. 2017;96(5):555-561. [46] HUYNH NC, EVERTS V, PAVASANT P, et al. Inhibition of Histone Deacetylases Enhances the Osteogenic Differentiation of Human Periodontal Ligament Cells. J Cell Biochem. 2016;117(6):1384-1395. [47] CHO YD, RYOO HM. Trans-differentiation via Epigenetics: A New Paradigm in the Bone Regeneration. J Bone Metab. 2018;25(1):9-13. [48] LOD S, JOHANSSON T, ABRAHAMSSON KH, et al. The influence of epigenetics in relation to oral health. Int J Dent Hyg. 2014;12(1):48-54. [49] SOMINENI HK, VENKATESWARAN S, KILARU V, et al. Blood-Derived DNA Methylation Signatures of Crohn’s Disease and Severity of Intestinal Inflammation. Gastroenterology. 2019;156(8):2254-2265. [50] ZHAO M, ZHOU Y, ZHU B, et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann Rheum Dis. 2016;75(11):1998-2006. [51] JIANG Y, FU J, DU J, et al. DNA methylation alterations and their potential influence on macrophage in periodontitis. Oral Dis. 2020 Sep 29. doi: 10.1111/odi.13654. [52] KOJIMA A, KOBAYASHI T, ITO S, et al. Tumor necrosis factor-alpha gene promoter methylation in Japanese adults with chronic periodontitis and rheumatoid arthritis. J Periodontal Res. 2016;51(3):350-358. [53] KURUSHIMA Y, TSAI PC, CASTILLO-FERNANDEZ J, et al. Epigenetic findings in periodontitis in UK twins: a cross-sectional study. Clin Epigenetics. 2019;11(1):27-27. [54] WEI Y, SHI M, ZHEN M, et al. Comparison of subgingival and buccal mucosa microbiome in chronic and aggressive periodontitis: a pilot study. Front Cell Infect Microbiol. 2019;9(1):53-53. [55] EIPEL M, MAYER F, ARENT T, et al. Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures. Aging (Albany NY). 2016;8(5):1034-1048. [56] LANGIE SAS, MOISSE M, DECLERCK K, et al. Salivary DNA Methylation Profiling: Aspects to Consider for Biomarker Identification. Basic Clin Pharmacol Toxicol. 2017;121 (Suppl 3):93-101. [57] DE FARIA AMORMINO SA, ARÃO TC, SARAIVA AM, et al. Hypermethylation and low transcription of TLR2 gene in chronic periodontitis. Hum Immunol. 2013;74(9):1231-1236. [58] HAN P,IVANOVSKI S. Effect of Saliva Collection Methods on the Detection of Periodontium-Related Genetic and Epigenetic Biomarkers—A Pilot Study. Int J Mol Sci. 2019;20(19):4729-4729. [59] BENAKANAKERE MR, FINOTI L, PALIOTO DB, et al. Epigenetics, Inflammation, and Periodontal Disease. Current Oral Health Reports. 2019;6(1):37-46. [60] IVANOV M, BARRAGAN I, INGELMAN-SUNDBERG M. Epigenetic mechanisms of importance for drug treatment. Trends Pharmacol Sci. 2014;35(8):384-396. [61] CANTLEY MD, ZANNETTINO ACW, BARTOLD PM, et al. Histone deacetylases (HDAC) in physiological and pathological bone remodelling. Bone. 2017;95(1):162-174. [62] MENG S, ZHANG L, TANG Y, et al. BET Inhibitor JQ1 Blocks Inflammation and Bone Destruction. J Dent Res. 2014;93(7):657-662. [63] KIM TI, HAN JE, JUNG HM, et al. Analysis of histone deacetylase inhibitor-induced responses in human periodontal ligament fibroblasts. Biotechnol Lett.2013; 35(1): 129-133. [64] CANTLEY MD, BARTOLD PM, MARINO V, et al. Histone deacetylase inhibitors and periodontal bone loss. J Periodontal Res. 2011;46(6): 697-703. [65] MAKSYLEWICZ A, BYSIEK A, LAGOSZ KB, et al. BET Bromodomain Inhibitors Suppress Inflammatory Activation of Gingival Fibroblasts and Epithelial Cells From Periodontitis Patients. Front Immunol. 2019; 10(12): 933-933. [66] SUFARU IG, BEIKIRCHER G, WEINHAEUSEL A, et al. Inhibitors of DNA methylation support TGF-β1-induced IL11 expression in gingival fibroblasts. J Periodontal Implant Sci. 2017;47(2):66-76. [67] TOMASELLI D, LUCIDI A, ROTILI D, et al. Epigenetic polypharmacology: A new frontier for epi-drug discovery. Med Res Rev. 2020;40(1):190-244. [68] DUVIC M, DIMOPOULOS M. The safety profile of vorinostat (suberoylanilide hydroxamic acid) in hematologic malignancies: A review of clinical studies. Cancer Treat Rev. 2016;43(1):58-66. [69] STATHIS A, BERTONI F. BET Proteins as Targets for Anticancer Treatment. Cancer Discov. 2018;8(1):24-36. [70] LARSSON L, DECKER AM, NIBALI L, et al. Regenerative Medicine for Periodontal and Peri-implant Diseases. J Dent Res. 2016;95(3):255-266. [71] BRACK AS, CONBOY MJ, ROY S, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007; 317(5839):807-810. [72] VISHAL M, AJEETHA R, KEERTHANA R, et al. Regulation of Runx2 by Histone Deacetylases in Bone. Curr Protein Pept Sci. 2016;17(4):343-351. [73] ZHANG D, LI Q, RAO L, et al. Effect of 5-Aza-2’-deoxycytidine on odontogenic differentiation of human dental pulp cells. J Endod. 2015; 41(5):640-645. [74] GOPINATHAN G, KOLOKYTHAS A, LUAN X, et al. Epigenetic marks define the lineage and differentiation potential of two distinct neural crest-derived intermediate odontogenic progenitor populations. Stem Cells Dev. 2013;22(12):1763-1778. [75] LIU Z, CHEN T, SUN W, et al. DNA demethylation rescues the impaired osteogenic differentiation ability of human periodontal ligament stem cells in high glucose. Scientific Reports. 2016;6(1):1-12. [76] HU X, ZHANG X, DAI L, et al. Histone deacetylase inhibitor trichostatin A promotes the osteogenic differentiation of rat adipose-derived stem cells by altering the epigenetic modifications on Runx2 promoter in a BMP signaling-dependent manner. Stem Cells Dev. 2013;22(2):248-255. [77] GE W, LIU Y, CHEN T, et al. The epigenetic promotion of osteogenic differentiation of human adipose-derived stem cells by the genetic and chemical blockade of histone demethylase LSD1. Biomaterials. 2014;35(23) 6015-6025. [78] DENG L, HONG H, ZHANG X, et al. Down-regulated lncRNA MEG3 promotes osteogenic differentiation of human dental follicle stem cells by epigenetically regulating Wnt pathway. Biochem Biophys Res Commun. 2018;503(3):2061-2067. [79] LI Z, GUO X, WU S. Epigenetic silencing of KLF2 by long non-coding RNA SNHG1 inhibits periodontal ligament stem cell osteogenesis differentiation. Stem Cell Res Ther. 2020;11(1):435-435. [80] NAIR A,TANG L. Influence of scaffold design on host immune and stem cell responses. Semin Immunol. 2017;29(1):62-71. [81] RABINEAU M, FLICK F, MATHIEU E, et al. Cell guidance into quiescent state through chromatin remodeling induced by elastic modulus of substrate. Biomaterials. 2015;37(2):144-155. [82] LV L, LIU Y, ZHANG P, et al. The nanoscale geometry of TiO2 nanotubes influences the osteogenic differentiation of human adipose-derived stem cells by modulating H3K4 trimethylation. Biomaterials. 2015; 39(3):193-205. [83] HA SW, JANG HL, NAM KT, et al. Nano-hydroxyapatite modulates osteoblast lineage commitment by stimulation of DNA methylation and regulation of gene expression. Biomaterials. 2015;65(1):32-42. [84] DU M, DUAN X, YANG P. Induced Pluripotent Stem Cells and Periodontal Regeneration. Curr Oral Health Rep. 2015;2(4):257-265. [85] JIA X, MIRON RJ, YIN C, et al. HnRNPL inhibits the osteogenic differentiation of PDLCs stimulated by SrCl(2) through repressing Setd2. J Cell Mol Med. 2019;23(4):2667-2677. [86] LORDEN ER, LEVINSON HM,LEONG KW. Integration of drug, protein, and gene delivery systems with regenerative medicine. Drug Deliv Transl Res. 2015;5(2):168-186. |
| [1] | 柴 乐, 吕建兰, 胡劲涛, 胡华辉, 许庆军, 余进伟, 全仁夫. 诱导急性脊髓损伤模型大鼠炎症反应信号通路的变化[J]. 中国组织工程研究, 2021, 25(8): 1218-1223. |
| [2] | 宋 珊, 胡方媛, 乔 军, 王 佳, 张升校, 李小峰. 基于生物信息学途径认识骨关节炎滑膜的生物学标志物[J]. 中国组织工程研究, 2021, 25(5): 785-790. |
| [3] | 邓桢翰, 黄 勇, 肖璐璐, 陈昱霖, 朱伟民, 陆 伟, 王大平. 骨形态发生蛋白在关节软骨再生过程中的作用与应用[J]. 中国组织工程研究, 2021, 25(5): 798-806. |
| [4] | 王玉姣, 刘 丹, 孙 嵩, 孙 勇. 改良型富血小板纤维蛋白复合双相磷酸钙可促进兔骨髓间充质干细胞的活性[J]. 中国组织工程研究, 2021, 25(4): 504-509. |
| [5] | 刘江锋. 纳米羟基磷灰石/聚酰胺66复合材料联合锁定钢板治疗股骨骨纤维异常增殖症[J]. 中国组织工程研究, 2021, 25(4): 542-547. |
| [6] | 肖 莎, 高承志, 周冬平. 三种骨替代材料修复即刻种植下颌后牙区周围骨缺损的比较[J]. 中国组织工程研究, 2021, 25(34): 5495-5500. |
| [7] | 刘日旭, 吕文波, 高文山. 磷酸钙骨水泥改性相关研究与临床应用[J]. 中国组织工程研究, 2021, 25(34): 5537-5543. |
| [8] | 范鑫超, 鲍文娟, 张 凯, 孙喜龙, 黄 腾, 高 博, 翟金帅, 周逸彬, 邱长茂, 李文毅, 李西成. D-二聚体、红细胞沉降率和C-反应蛋白在髋、膝关节置换后下肢深静脉血栓形成中的诊断价值#br#[J]. 中国组织工程研究, 2021, 25(33): 5324-5328. |
| [9] | 伍伟挺, 黎润光, 曹生鲁, 梁双武. 过度周期性机械应力刺激可引起软骨细胞炎症反应及凋亡[J]. 中国组织工程研究, 2021, 25(29): 4608-4613. |
| [10] | 陈 赵, 莫雨晴, 唐宏宇. 电针干预去卵巢骨质疏松模型大鼠骨组织Runx2启动子甲基化水平的变化[J]. 中国组织工程研究, 2021, 25(29): 4644-4649. |
| [11] | 贺生才, 蒋成丹, 荆纯祥, 罗敏怡, 马兴发, 李嘉洲, 潘华山. 滑膜液α防御素和C-反应蛋白在关节假体周围感染诊断中的价值[J]. 中国组织工程研究, 2021, 25(27): 4344-4347. |
| [12] | 覃健芳, 王 欢, 吴冰冰, 马小京. GOLM1基因敲除对单侧输尿管梗阻小鼠肾纤维化的影响[J]. 中国组织工程研究, 2021, 25(26): 4162-4167. |
| [13] | 彭竑程, 华 臻, 王建伟. MiR-21——骨再生及多种骨疾病的重要调控因子[J]. 中国组织工程研究, 2021, 25(26): 4198-4203. |
| [14] | 阮光萍, 姚 翔, 刘高米洋, 蔡学敏, 李自安, 庞荣清, 王金祥, 潘兴华. 脐带间充质干细胞移植治疗树鼩创伤性全身炎症反应综合征[J]. 中国组织工程研究, 2021, 25(25): 3994-4000. |
| [15] | 程崇杰, 闫 延, 张启栋, 郭万首. D-二聚体在诊断假体周围感染中的应用价值及准确性:系统评价与荟萃分析[J]. 中国组织工程研究, 2021, 25(24): 3921-3928. |
1.2 检索方法
纳入标准:①有关表观遗传学的概述及作用机制的文献;②有关牙周疾病与表观遗传研究的文献;③有关牙周细胞调节骨骼再生的表观遗传研究文献;④有关表观遗传学在牙周领域的临床应用研究文献;⑤有关牙周组织工程研究的文献。
排除标准:重复性研究及关系不密切的文献。
质量评估:通过阅读文题和摘要进行初筛,再经过进一步查阅全文及文章撰写时再次筛选取舍,最终保留86篇相关文献,均为与此次综述目的密切相关的、近5年的实验研究和临床研究。文献检索流程图见图1。

文题释义:
表观遗传学:是研究基因的核苷酸序列不发生改变的情况下,基因表达的可遗传变化的一门遗传学分支学科。表观遗传的现象很多,已知的有DNA甲基化(DNA methylation)、基因组印记(genomic imprinting)、母体效应(maternal effects)、基因沉默(gene silencing)、核仁显性、休眠转座子激活和RNA编辑(RNA editing)等。
DNA甲基化(DNA methylation):为DNA化学修饰的一种形式,能够在不改变DNA序列的前提下改变遗传表现。所谓DNA甲基化是指在DNA甲基化转移酶的作用下,在基因组CpG二核苷酸的胞嘧啶5’碳位共价键结合一个甲基基团。大量研究表明,DNA甲基化能引起染色质结构、DNA构象、DNA稳定性及DNA与蛋白质相互作用方式的改变,从而控制基因表达。
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||