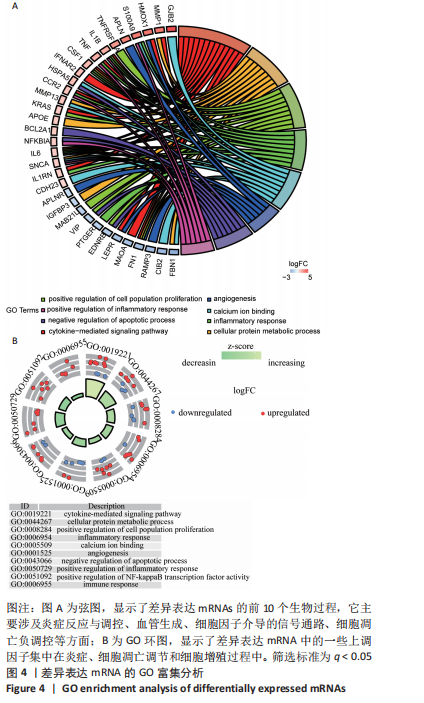
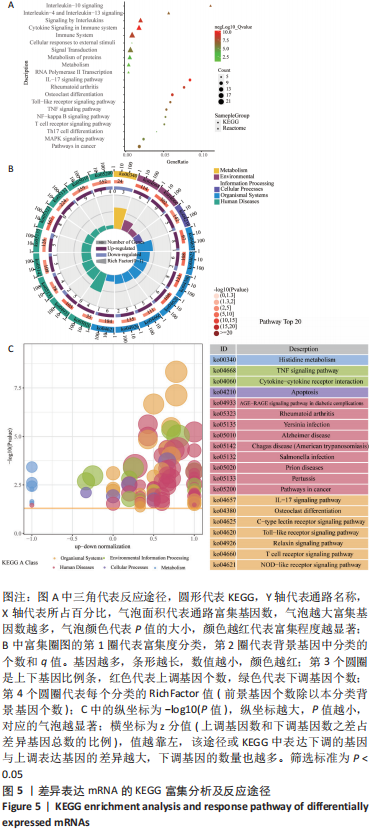
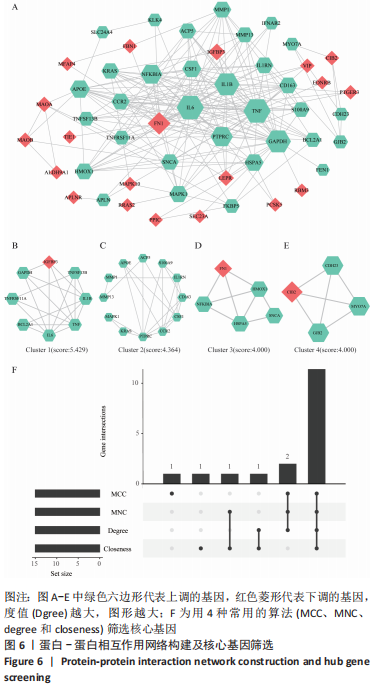
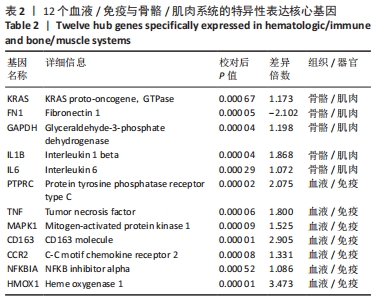
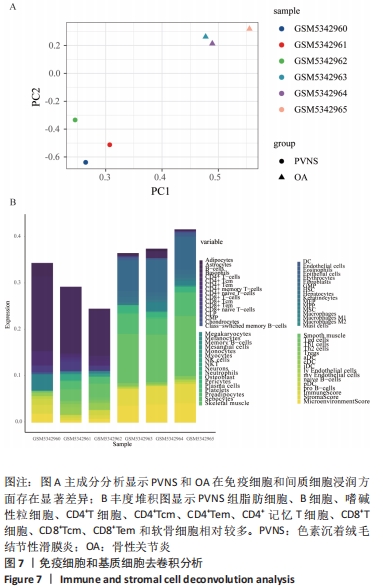
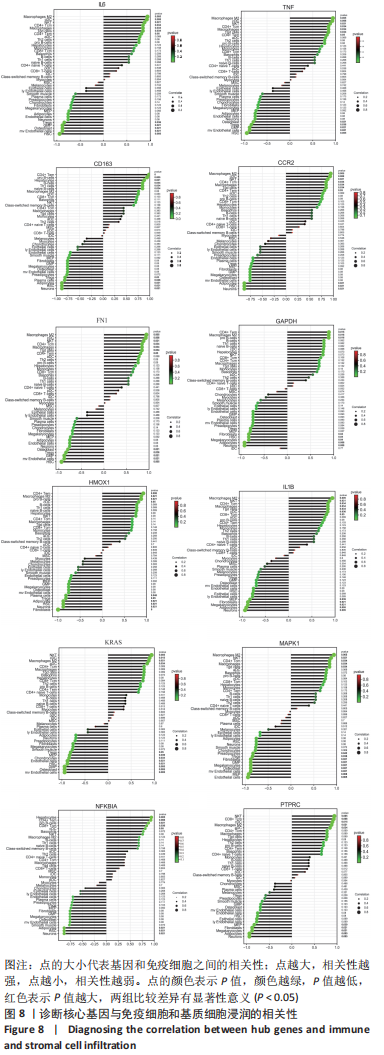
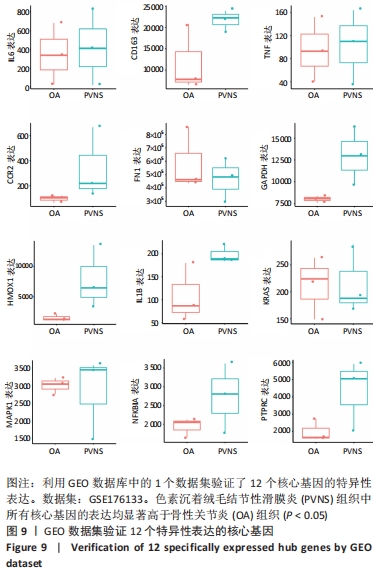
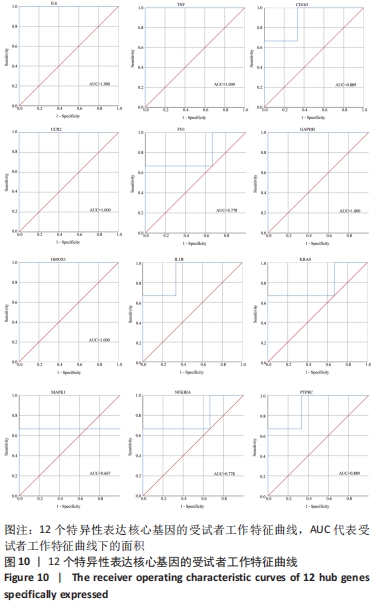
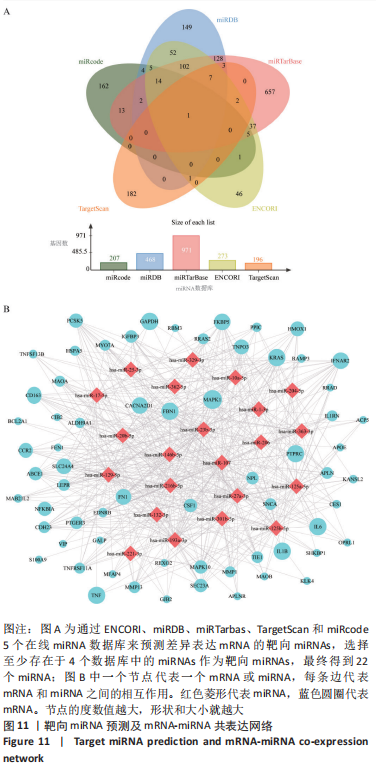
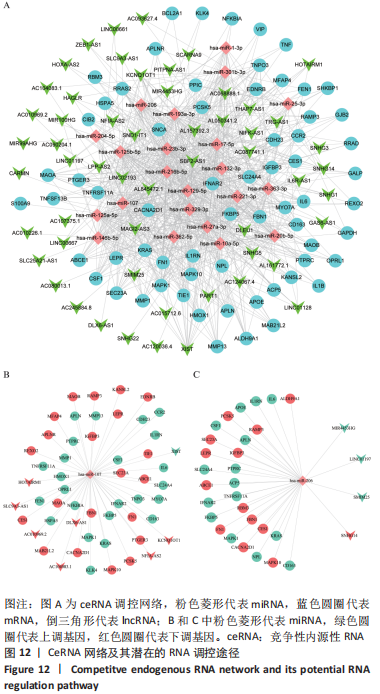
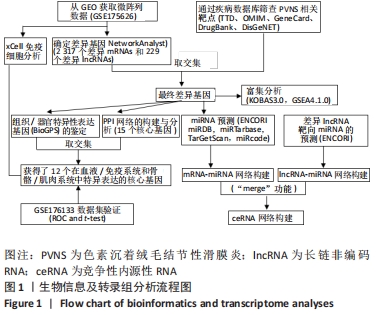
[1] TYLER WK, VIDAL AF, WILLIAMS RJ, et al. Pigmented villonodular synovitis. J Am Acad Orthop Surg. 2006;14(6):376-385.
[2] GEIGER EV, REIZE P, RUDERT M, et al. Pigmented villonodular synovitis. MMW Fortschr Med. 2006;148(6):40-41.
[3] COURT S, NISSEN MJ, GABAY C. Pigmented villonodular synovitis. Rev Med Suisse. 2014;10(421):609-610.
[4] FAŁEK A, NIEMUNIS-SAWICKA J, WRONA K, et al. Pigmented villonodular synovitis. Folia Med Cracov. 2018;58(4):93-104.
[5] XIE GP, JIANG N, LIANG CX, et al. Pigmented villonodular synovitis: a retrospective multicenter study of 237 cases. PLoS One. 2015;10(3): e0121451.
[6] FANG Y, ZHANG Q. Recurrence of pigmented villonodular synovitis of the knee: A case report with review of literature on the risk factors causing recurrence. Medicine (Baltimore). 2020;99(16):e19856.
[7] DÜRR HR, CAPELLEN CF, KLEIN A, et al. The effects of radiosynoviorthesis in pigmented villonodular synovitis of the knee. Arch Orthop Trauma Surg. 2019;139(5):623-627.
[8] MOLLON B, LEE A, BUSSE JW, et al. The effect of surgical synovectomy and radiotherapy on the rate of recurrence of pigmented villonodular synovitis of the knee: an individual patient meta-analysis. Bone Joint J. 2015;97-B(4):550-557.
[9] FECEK C, CARTER KR. Pigmented Villonodular Synovitis//StatPearls. Treasure Island (FL): StatPearls Publishing, 2021.
[10] GOUIN F, NOAILLES T. Localized and diffuse forms of tenosynovial giant cell tumor (formerly giant cell tumor of the tendon sheath and pigmented villonodular synovitis). Orthop Traumatol Surg Res. 2017; 103(1S):S91-S97.
[11] EFRIMA B, SAFRAN N, AMAR E, et al. Simultaneous pigmented villonodular synovitis and synovial chondromatosis of the hip: case report. J Hip Preserv Surg. 2018;5(4):443-447.
[12] DYADYK OO, HRYHOROVSKA AV. Clinical and morphological correlations and histopathology of joint damage in patients with diffuse-type tenosynovial giant cell tumor. Wiad Lek. 2019;72(12 cz 1):2269-2276.
[13] JAFFE HL, LICHTENSTEIN L, SUTRO CJ. Pigmented villonodular synovitis, bursitis and tenosynovitis: A discussion of the synovial and bursal equivalents of the tenosynovial lesion commonly denoted as xanthoma, xanthogranuloma, giant cell tumor or myeloplaxoma of tendon sheath with some consideration of this tendon sheath lesionitself. Arch Pathol 1941;31:731-765.
[14] BERGER I, WECKAUF H, HELMCHEN B, et al. Rheumatoid arthritis and pigmented villonodular synovitis: comparative analysis of cell polyploidy, cell cycle phases and expression of macrophage and fibroblast markers in proliferating synovial cells. Histopathology. 2005; 46(5):490-497.
[15] O’KEEFE RJ, ROSIER RN, TEOT LA, et al. Cytokine and matrix metalloproteinase expression in pigmented villonodular synovitis may mediate bone and cartilage destruction. Iowa Orthop J. 1998;18:26-34.
[16] KROOT EJ, KRAAN MC, SMEETS TJ, et al. Tumour necrosis factor alpha blockade in treatment resistant pigmented villonodular synovitis. Ann Rheum Dis. 2005;64(3):497-499.
[17] RITCHIE ME, PHIPSON B, WU D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
[18] CAI W, LI H, ZHANG Y, et al. Identification of key biomarkers and immune infiltration in the synovial tissue of osteoarthritis by bioinformatics analysis. PeerJ. 2020;8:e8390.
[19] TAO Z, SHI A, LI R, et al. Microarray bioinformatics in cancer- a review. J BUON. 2017;22(4):838-843.
[20] SALMENA L, POLISENO L, TAY Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146(3):353-358.
[21] BARRETT T, WILHITE SE, LEDOUX P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013; 41(Database issue):D991-D995.
[22] ZHOU G, SOUFAN O, EWALD J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234-W241.
[23] WANG Y, ZHANG S, LI F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031-D1041.
[24] AMBERGER JS, BOCCHINI CA, SCHIETTECATTE F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789-D798.
[25] STELZER G, ROSEN N, PLASCHKES I, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1-1.30.33.
[26] WISHART DS, FEUNANG YD, GUO AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46: D1074-D1082.
[27] PIÑERO J, BRAVO À, QUERALT-ROSINACH N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833-D839.
[28] WU C, JIN X, TSUENG G, et al. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016; 44(D1):D313-D316.
[29] SZKLARCZYK D, GABLE AL, LYON D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613.
[30] BADER GD, HOGUE CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2.
[31] CHIN CH, CHEN SH, WU HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4(Suppl 4):S11.
[32] SUBRAMANIAN A, KUEHN H, GOULD J, et al. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007; 23(23):3251-3253.
[33] BU D, LUO H, HUO P, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49(W1):W317-W325.
[34] ARAN D, HU Z, BUTTE AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220.
[35] JOLLIFFE IT, CADIMA J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374 (2065):20150202.
[36] LI JH, LIU S, ZHOU H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92-D97.
[37] CAO R, LÓPEZ-DE-ULLIBARRI I. ROC Curves for the Statistical Analysis of Microarray Data. Methods Mol Biol. 2019;1986:245-253.
[38] NI Z, SHANG X, TANG G, et al. Expression of miR-206 in Human Knee Articular Chondrocytes and Effects of miR-206 on Proliferation and Apoptosis of Articular Chondrocytes. Am J Med Sci. 2018;355(3):240-246.
[39] ZHANG H, WANG J, REN T, et al. Bone marrow mesenchymal stem cell-derived exosomal miR-206 inhibits osteosarcoma progression by targeting TRA2B. Cancer Lett. 2020;490:54-65.
[40] ZHANG Z, WU S, MUHAMMAD S, et al. miR-103/107 promote ER stress-mediated apoptosis via targeting the Wnt3a/β-catenin/ATF6 pathway in preadipocytes. J Lipid Res. 2018;59(5):843-853.
[41] ZHAO X, LI H, WANG L. MicroRNA-107 regulates autophagy and apoptosis of osteoarthritis chondrocytes by targeting TRAF3. Int Immunopharmacol. 2019;71:181-187.
[42] SHEKHAR A, SINGH S, PATIL SS, et al. Osteochondral Lesion in Diffuse Pigmented Villonodular Synovitis of the Knee. Knee Surg Relat Res. 2019;31(1):67-71.
[43] FECEK C, CARTER KR. Pigmented Villonodular Synovitis. In: StatPearls. Treasure Island (FL): StatPearls Publishing, 2022.
[44] SUN R, TIAN Z, KULKARNI S, et al. IL-6 prevents T cell-mediated hepatitis via inhibition of NKT cells in CD4+ T cell- and STAT3-dependent manners. J Immunol. 2004;172(9):5648-5655.
[45] NAGASHIMA H, ISHII N, SO T. Regulation of Interleukin-6 Receptor Signaling by TNF Receptor-Associated Factor 2 and 5 During Differentiation of Inflammatory CD4+ T Cells. Front Immunol. 2018;9: 1986.
[46] POVOLERI GAM, LALNUNHLIMI S, STEEL KJA, et al. Anti-TNF treatment negatively regulates human CD4+ T-cell activation and maturation in vitro, but does not confer an anergic or suppressive phenotype. Eur J Immunol. 2020;50(3):445-458.
[47] ZHOU WH, DU WD, LI YF, et al. The Overexpression of Fibronectin 1 Promotes Cancer Progression and Associated with M2 Macrophages Polarization in Head and Neck Squamous Cell Carcinoma Patients. Int J Gen Med. 2022;15:5027-5042.
[48] NEES TA, ROSSHIRT N, REINER T, et al. Inflammation and osteoarthritis-related pain. Schmerz. 2019;33(1):4-12.
[49] JAMAL J, ROEBUCK MM, LEE SY, et al. Modulation of the mechanical responses of synovial fibroblasts by osteoarthritis-associated inflammatory stressors. Int J Biochem Cell Biol. 2020;126:105800.
[50] WALLACH D. The cybernetics of TNF: Old views and newer ones. Semin Cell Dev Biol. 2016;50:105-114.
[51] GREISEN SR, MOLLER HJ, STENGAARD-PEDERSEN K, et al. Soluble macrophage-derived CD163 is a marker of disease activity and progression in early rheumatoid arthritis. Clin Exp Rheumatol. 2011; 29(4):689-692.
[52] BERGER I, EHEMANN V, HELMCHEN B, et al. Comparative analysis of cell populations involved in the proliferative and inflammatory processes in diffuse and localised pigmented villonodular synovitis. Histol Histopathol. 2004;19(3):687-692.
[53] OHASHI Y, UCHIDA K, FUKUSHIMA K, et al. Correlation between CD163 expression and resting pain in patients with hip osteoarthritis: Possible contribution of CD163+ monocytes/macrophages to pain pathogenesis. J Orthop Res. 2022;40(6):1365-1374.
[54] WU HZ, XIAO JQ, XIAO SS, et al. KRAS: A Promising Therapeutic Target for Cancer Treatment. Curr Top Med Chem. 2019;19(23):2081-2097.
[55] HAMARSHEH S, GROß O, BRUMMER T, et al. Immune modulatory effects of oncogenic KRAS in cancer. Nat Commun. 2020;11(1):5439.
[56] WU Z, SHOU L, WANG J, et al. Identification of the key gene and pathways associated with osteoarthritis via single-cell RNA sequencing on synovial fibroblasts. Medicine (Baltimore). 2020;99(33):e21707.
[57] YANG S, OHE R, AUNG NY, et al. Comparative study of HO-1 expressing synovial lining cells between RA and OA. Mod Rheumatol. 2021;31(1): 133-140.
[58] SANADA Y, TAN SJO, ADACHI N, et al. Pharmacological Targeting of Heme Oxygenase-1 in Osteoarthritis. Antioxidants (Basel). 2021; 10(3):419.
[59] ISHIHARA S, OBEIDAT AM, WOKOSIN DL, et al. The role of intra-articular neuronal CCR2 receptors in knee joint pain associated with experimental osteoarthritis in mice. Arthritis Res Ther. 2021;23(1):103.
[60] WANG Z, WANG B, ZHANG J, et al. Chemokine (C-C Motif) Ligand 2/Chemokine Receptor 2 (CCR2) Axis Blockade to Delay Chondrocyte Hypertrophy as a Therapeutic Strategy for Osteoarthritis. Med Sci Monit. 2021;27:e930053.
[61] DECLERCQ HA, FORSYTH RG, VERBRUGGEN A, et al. CD34 and SMA expression of superficial zone cells in the normal and pathological human meniscus. J Orthop Res. 2012;30(5):800-808.
[62] KUBSIK-GIDLEWSKA A, KLUPIŃSKI K, KROCHMALSKI M, et al. CD34+ Stem Cell Treatment for Knee Osteoarthritis: A Treatment and Rehabilitation Algorithm. J Rehabil Med Clin Commun. 2018;3:1000012.
[63] WANG B, LI J, TIAN F. Downregulation of lncRNA SNHG14 attenuates osteoarthritis by inhibiting FSTL-1 mediated NLRP3 and TLR4/NF-κB pathway through miR-124-3p. Life Sci. 2021;270:119143.
[64] LIU Y, LIU K, TANG C, et al. Long non-coding RNA XIST contributes to osteoarthritis progression via miR-149-5p/DNMT3A axis. Biomed Pharmacother. 2020;128:110349.
|