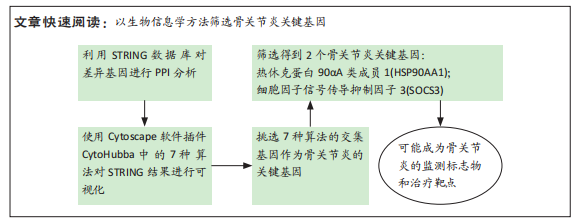
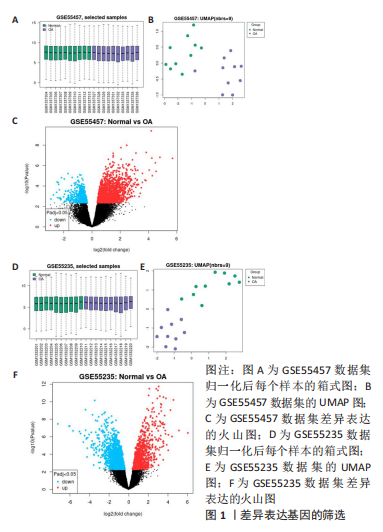
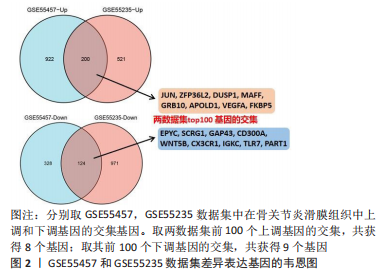
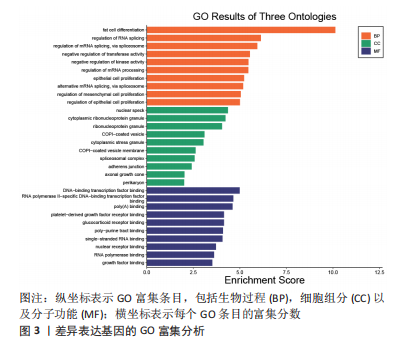
[1] MOTTA F, BARONE E, SICA A, et al. Inflammaging and Osteoarthritis. Clin Rev Allergy Immunol.2022 Jun 18. doi: 10.1007/s12016-022-08941-1.
[2] MA L, CHHETRI JK, ZHANG L, et al. Cross-sectional study examining the status of intrinsic capacity decline in community-dwelling older adults in China: prevalence, associated factors and implications for clinical care. BMJ Open. 2021;11(1):e043062.
[3] LO J, CHAN L, FLYNN S. A Systematic Review of the Incidence, Prevalence, Costs, and Activity and Work Limitations of Amputation, Osteoarthritis, Rheumatoid Arthritis, Back Pain, Multiple Sclerosis, Spinal Cord Injury, Stroke, and Traumatic Brain Injury in the United States: A 2019 Update. Arch Phys Med Rehabil. 2021;102(1):115-131.
[4] HUNTER D J, MARCH L, CHEW M. Osteoarthritis in 2020 and beyond: a Lancet Commission. Lancet. 2020;396(10264):1711-1712.
[5] GBD 2019 Diseases and Injuries Collaborators.Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396(10258):1204-1222.
[6] HUNTER DJ, BIERMA-ZEINSTRA S. Osteoarthritis. Lancet. 2019;393(10182):1745-1759.
[7] 苏剑清, 孙波, 丁韵蓉, 等. 基于“损伤-修复-再损伤”方法制备兔膝骨关节炎滑膜炎模型[J]. 中国组织工程研究,2022,26(23):3738-3743.
[8] SANCHEZ-LOPEZ E, CORAS R, TORRES A, et al. Synovial inflammation in osteoarthritis progression. Nat Rev Rheumatol. 2022;18(5):258-275.
[9] ZHANG L, XING R, HUANG Z, et al. Synovial Fibrosis Involvement in Osteoarthritis. Front Med (Lausanne). 2021;8:684389.
[10] KULKARNI P, MARTSON A, VIDYA R, et al. Pathophysiological landscape of osteoarthritis. Adv Clin Chem. 2021;100:37-90.
[11] DE REZENDE MU, DE CAMPOS GC. Is osteoarthritis a mechanical or inflammatory disease?. Rev Bras Ortop. 2013;48(6):471-474.
[12] SMITH MM, CAKE MA, GHOSH P, et al. Significant synovial pathology in a meniscectomy model of osteoarthritis: modification by intra-articular hyaluronan therapy. Rheumatology (Oxford). 2008;47(8): 1172-1178.
[13] 张虎林, 王亮, 喻琳, 等. 滑膜巨噬细胞在骨性关节炎发病机制中的作用及其潜在性应用研究进展[J]. 中国骨质疏松杂志,2022,28(4):590-595.
[14] XIE J, HUANG Z, YU X, et al. Clinical implications of macrophage dysfunction in the development of osteoarthritis of the knee. Cytokine Growth Factor Rev. 2019;46:36-44.
[15] ALAHDAL M, ZHANG H, HUANG R, et al. Potential efficacy of dendritic cell immunomodulation in the treatment of osteoarthritis. Rheumatology (Oxford). 2021;60(2):507-517.
[16] MATHIESSEN A, CONAGHAN PG. Synovitis in osteoarthritis: current understanding with therapeutic implications. Arthritis Res Ther. 2017;19(1):18.
[17] MURPHY CA, GARG AK, SILVA-CORREIA J, et al. The Meniscus in Normal and Osteoarthritic Tissues: Facing the Structure Property Challenges and Current Treatment Trends. Annu Rev Biomed Eng. 2019;21:495-521.
[18] ZHAO Y, LV J, ZHANG H, et al. Gene Expression Profiles Analyzed Using Integrating RNA Sequencing, and Microarray Reveals Increased Inflammatory Response, Proliferation, and Osteoclastogenesis in Pigmented Villonodular Synovitis. Front Immunol. 2021;12:665442.
[19] MAKSYMOWYCH WP, RUSSELL AS, CHIU P, et al. Targeting tumour necrosis factor alleviates signs and symptoms of inflammatory osteoarthritis of the knee. Arthritis Res Ther. 2012;14(5):R206.
[20] ZHANG L, CHEN X, CAI P, et al. Reprogramming Mitochondrial Metabolism in Synovial Macrophages of Early Osteoarthritis by a Camouflaged Meta-Defensome. Adv Mater. 2022;34(30):e2202715.
[21] GUI T, LUO L, CHHAY B, et al. Superoxide dismutase-loaded porous polymersomes as highly efficient antioxidant nanoparticles targeting synovium for osteoarthritis therapy. Biomaterials. 2022;283:121437.
[22] LI M, HAN H, CHEN L, et al. Platelet-rich plasma contributes to chondroprotection by repairing mitochondrial function via AMPK/NF-κB signaling in osteoarthritic chondrocytes. Tissue Cell. 2022;77:101830.
[23] CAI Y, WANG Z, LIAO B, et al. Anti-inflammatory and chondroprotective effects of platelet-derived growth factor-BB on osteoarthritis rat models. J Gerontol A Biol Sci Med Sci. 2022;glac118.
[24] MALAISE O, RELIC B, CHARLIER E, et al. Glucocorticoid-induced leucine zipper (GILZ) is involved in glucocorticoid-induced and mineralocorticoid-induced leptin production by osteoarthritis synovial fibroblasts. Arthritis Res Ther. 2016; 18(1):219.
[25] SAVVIDOU O, MILONAKI M, GOUMENOS S, et al. Glucocorticoid signaling and osteoarthritis. Mol Cell Endocrinol. 2019;480:153-166.
[26] ZHANG Y, PIZZUTE T, PEI M. A review of crosstalk between MAPK and Wnt signals and its impact on cartilage regeneration. Cell Tissue Res. 2014;358(3):633-649.
[27] SCHETT G, KLEYER A, PERRICONE C, et al. Diabetes is an independent predictor for severe osteoarthritis: results from a longitudinal cohort study Diabetes Care. 2013;36(2):403-409.
[28] HAMADA D, MAYNARD R, SCHOTT E, et al. Suppressive Effects of Insulin on Tumor Necrosis Factor-Dependent Early Osteoarthritic Changes Associated With Obesity and Type 2 Diabetes Mellitus. Arthritis Rheumatol. 2016;68(6):1392-1402.
[29] GRIFFIN TM, HUFFMAN KM. Editorial: Insulin Resistance: Releasing the Brakes on Synovial Inflammation and Osteoarthritis?. Arthritis Rheumatol. 2016;68(6):1330-1333.
[30] TSAI CH, CHIANG YC, CHEN HT, et al. High glucose induces vascular endothelial growth factor production in human synovial fibroblasts through reactive oxygen species generation. Biochim Biophys Acta. 2013;1830(3):2649-2658.
[31] WANG Z, QI G, LI Z, et al. Effects of urolithin A on osteoclast differentiation induced by receptor activator of nuclear factor-κB ligand via bone morphogenic protein 2. Bioengineered. 2022;13(3):5064-5078.
[32] KIM GM, PARK H, LEE SY. Roles of osteoclast-associated receptor in rheumatoid arthritis and osteoarthritis. Joint Bone Spine. 2022;89(5):105400.
[33] XU Y, XUE S, ZHANG T, et al. Toddalolactone protects against osteoarthritis by ameliorating chondrocyte inflammation and suppressing osteoclastogenesis. Chin Med. 2022;17(1):18.
[34] JIANG S, ZHANG C, LU Y, et al. The molecular mechanism research of cartilage calcification induced by osteoarthritis. Bioengineered. 2022;13(5):13082-13088.
[35] CUNNINGHAM CC, CORR EM, MCCARTHY GM, et al. Intra-articular basic calcium phosphate and monosodium urate crystals inhibit anti-osteoclastogenic cytokine signalling. Osteoarthritis Cartilage. 2016;24(12):2141-2152.
[36] TAKAHASHI K, KUBO T, ARAI Y, et al. Localization of heat shock protein in osteoarthritic cartilage. Scand J Rheumatol. 1997;26(5):368-375.
[37] NGARMUKOS S, SCARAMUZZA S, THEERAWATTANAPONG N, et al. Circulating and Synovial Fluid Heat Shock Protein 70 Are Correlated with Severity in Knee Osteoarthritis. Cartilage. 2020;11(3):323-328.
[38] SON YO, KIM HE, CHOI WS, et al. RNA-binding protein ZFP36L1 regulates osteoarthritis by modulating members of the heat shock protein 70 family. Nat Commun. 2019;10(1):77.
[39] GUI T, HE BS, GAN Q, et al. Enhanced SOCS3 in osteoarthiritis may limit both proliferation and inflammation. Biotech Histochem. 2017;92(2):107-114. |