Chinese Journal of Tissue Engineering Research ›› 2023, Vol. 27 ›› Issue (33): 5370-5371.doi: 10.12307/2023.741
Previous Articles Next Articles
Application of single-cell RNA sequencing in intervertebral disc degeneration
Wu Hao1, Ye Dongping1, 2
- 1Guizhou Medical University, Guiyang 550025, Guizhou Province, China; 2Guangzhou Red Cross Hospital, Guangzhou 510220, Guangdong Province, China
-
Received:2022-10-10Accepted:2022-12-09Online:2023-11-28Published:2023-03-30 -
Contact:Ye Dongping, MD, Chief physician, Master’s supervisor, Guizhou Medical University, Guiyang 550025, Guizhou Province, China; Guangzhou Red Cross Hospital, Guangzhou 510220, Guangdong Province, China -
About author:Wu Hao, Master candidate, Physician, Guizhou Medical University, Guiyang 550025, Guizhou Province, China -
Supported by:Guangzhou Science and Technology Planning Project, No. 202102010111 (to YDP)
CLC Number:
Cite this article
Wu Hao, Ye Dongping. Application of single-cell RNA sequencing in intervertebral disc degeneration[J]. Chinese Journal of Tissue Engineering Research, 2023, 27(33): 5370-5371.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
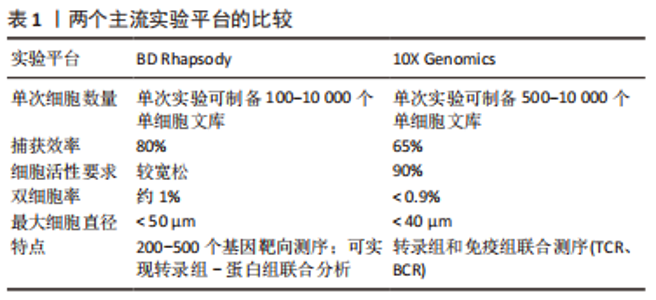
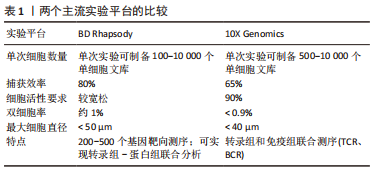
2.1 单细胞转录组测序技术 单细胞转录组测序技术是在单个细胞水平对mRNA进行高通量测序的一项新技术,原理是对单个细胞中微量的mRNA进行扩增后再进行高通量测序。单细胞转录组测序技术能够有效解决批量转录组测序技术掩盖的组织内部的转录组学异质性难题,有助于发现未被认知的微量细胞类型,并深入认知细胞生长过程中的表达调控机制。目前单细胞转录组测序已被广泛应用于肿瘤异质性[11]、免疫微环境[12]、生殖发育[13]、神经生长[14]、细胞分化等领域[15]。越来越多的学者应用单细胞转录组测序技术研究椎间盘退行性变,认识髓核、纤维环复杂的异质性,更进一步了解椎间盘组织内部丰富的分化与变性机制以及转录组学层面的信息,为以后从组织工程学、再生医学等角度探究椎间盘退行性变的治疗手段提供了可能。 2.1.1 单细胞转录组测序技术的步骤 单细胞转录组测序的主要流程包括组织的消化、单细胞的捕获、高通量测序、上游数据分析和下游数据分析等[16],不同平台的步骤大同小异。由于测序平台的成本高昂以及需要专业培训人员进行操作,单细胞转录组测序技术一般由商业化的服务公司提供。大部分情况下,研究者们仅需要选择合适的技术平台并完成头部和尾部的任务即可,包括组织的取样以及下游数据的分析。 源自Drop-Seq技术的10X Genomics与源于Cyto-Seq技术的BD Rhapsody平台每次实验可以捕获5 000-10 000个细胞[17-18],细胞捕获率大于65%,有测序通量高、商业化程度较高、单位成本较其他测序平台更低、适用范围更广等特点,两种平台数据的详细对比见表1。目前大部分已经报道的研究是基于这2种测序平台[19]。"
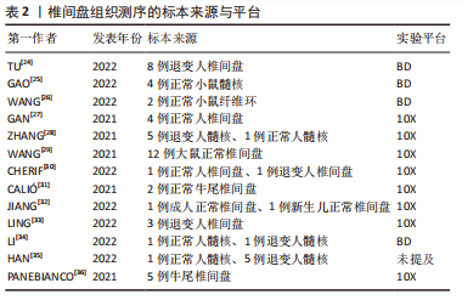
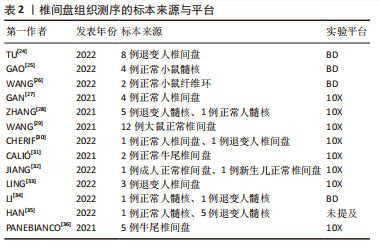
2.1.2 椎间盘组织的获取 大鼠、小鼠是最易获取的实验动物,并且可以对其进行臂丛神经根切断术以及远端骨骼和肌肉组织截肢手术以诱导其双足行走来建立椎间盘退变模型[20],所以大鼠、小鼠的椎间盘退变模型是研究人员使用最多的动物模型。由于牛尾肌肉可保持其椎间盘承受与人椎间盘相近的压力,人椎间盘与牛尾椎间盘有相近的尺寸和解剖学结构,牛尾椎间盘被视为研究腰椎间盘的生物力学模型[21],也有部分研究人员使用此模型获取退变椎间盘组织。人的正常椎间盘组织主要来源于尸体或者脊柱骨折手术获取的标本;人的退变椎间盘组织来源于腰椎融合术中摘除的标本。大部分研究者都根据MRI结果对椎间盘退变程度进行了Pfirrmann评级或者改良Pfirrmann评级[22-23],并以此来将椎间盘标本定义为轻、中、重等退变严重程度。表2列出目前椎间盘单细胞转录组测序研究所使用的标本来源。"
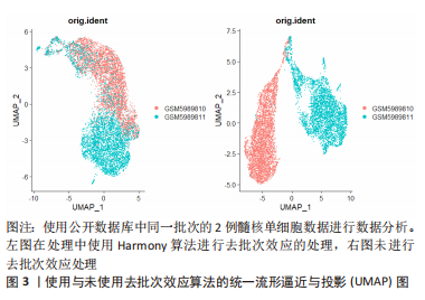
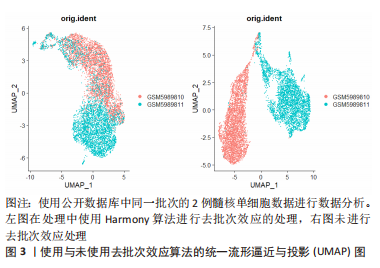
2.1.3 下游数据的分析 用于单细胞转录组测序下游数据分析的工具是由多种编程语言编写的,最主要的是R和Python。基于R语言的Seurat、Scater或Scanpy等常用软件包提供了集成环境来开发编写代码并包含大量分析工具[37–39]。但是,为了便于软件的更新升级,这些平台限制了它们只能使用各自的编程语言开发工具来使用。一台性能较高的服务器或者性能相当的个人电脑即可运行这些软件进行数据的下游分析。 2.1.4 单细胞转录组测序的局限性 受限于单细胞转录组测序技术的繁多操作步骤,下游数据的批次效应异常明显。测序反应过程中条件的设置、操作人员的操作习惯以及其他客观因素的影响,都会造成显著的批次差异,如果组织在不同批次进行转录组测序,批次间的差异可能远大于组织内细胞间差异,导致后续分析不能还原细胞间的真实差异[40]。为了解决这个问题,专业的生信软件开发人员开发出了处理批次效应的软件,例如Harmony[41]、LIGER[42]、MNN[43]、DESC等[44],这些软件可以很好的处理不同类型组织、相同平台不同批次间的批次差异[45]。但过度的或者不合适的批次差异处理会消除研究者期盼得到的真实的细胞转录组差异。因此,需要研究者选择合适的方法和适度的参数来处理数据的批次效应[46]。图3所示,使用同一批次的2例髓核单细胞数据进行分析,其中左图为经过去批次效应后的统一流形逼近与投影(Uniform Manifold Approximation and Projection,UMAP)图,右图为没有经过去批次效应处理的UMAP图,可见未经过去批次效应处理的UMAP图中,两组相同组织的细胞被划分为了2个亚群,而经过去批次效应算法处理后的UMAP图中,两组细胞被拟合在一起。"
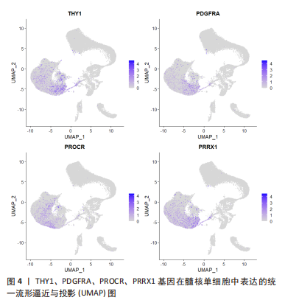
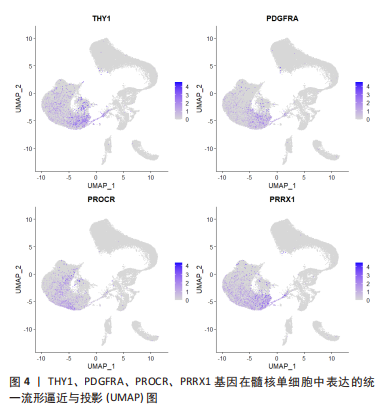
2.2 单细胞转录组测序在椎间盘退行性变中的应用 2.2.1 椎间盘的异质性 椎间盘主要由3个部分组成:中间的髓核、周围的纤维环以及与软骨相邻的终板。既往的研究已经通过批量RNA测序揭示了髓核、纤维环在转录组层面上的差别[47],单细胞测序对此进行了更深入详细的探究。 WANG等[29]对12只大鼠的椎间盘包括髓核、纤维环进行了单细胞转录组测序,验证了经典的髓核、纤维环组织标志基因:髓核细胞特异表达CD24、Anxa3和Krt19,纤维环细胞特异表达Acan、Col1a1和Col2a1等;确定了一组特异性更强的标记基因:髓核细胞表达Krt7、Prrg4、Akap12、Cxcl3、Rab38、Illrl1等,纤维环细胞表达Fibin、Myoc、Igfbp5、Cilp、Tnmd、Lum等。GAN等[27]对5名健康人的椎间盘进行测序后发现了7种不同类型的细胞:脊索细胞、基质细胞、周皮细胞、内皮细胞、髓核祖细胞、血细胞等,椎间盘的3个部分分别包含不同比例的以上7种类型细胞。同样的,PANEBIANCO等[36]对来自3个牛尾的5个椎间盘进行了测序,也发现了仅存在于髓核组织中的脊索细胞。 单细胞测序更容易区分出不同组织在功能上的差异。WANG等[29]研究报道:髓核的功能与细胞外基质有关,而纤维环的功能与应激反应和纤维化疾病有关。CALIó等[31]对牛尾椎间盘的研究也表明:相比较纤维环,髓核显著富集胶原代谢过程,但与上述研究相悖的是髓核有对应激反应的DNA转录调控的功能。GAN等[27]的研究更细致地对每个亚群进行了功能富集研究,根据每种组织包含亚群比例不同,推断出3种组织不同的功能:主要存在于纤维环和终板的软骨亚群包含显著的氧化磷酸化代谢模式,而进行糖酵解的亚群出现在无血管缺氧环境的髓核组织中。 2.2.2 髓核组织的异质性 髓核的变性是椎间盘退变的主要原因。既往的研究无法详细探究髓核的异质性,为了解决这个问题,所有的椎间盘单细胞测序研究都进行了髓核组织的异质性分析。 ZHANG等[28]对5例退变椎间盘的髓核和1例正常椎间盘的髓核进行了转录组测序,得出椎间盘髓核组织中包含的3个差异显著的细胞亚群:在所有髓核细胞中占比99.33%的软骨细胞,占所有细胞0.57%的巨噬细胞,约占0.10%的内皮细胞。再次对软骨细胞划分得到了具有多分化能力以保持纤维软骨细胞发育的纤维软骨细胞祖细胞;具有自我更新、有沿多个谱系分化的能力、表达干细胞相关表面标志物并且在正常髓核细胞中占比较多的软骨祖细胞;有助于调节软骨细胞代谢活动的稳态平衡以响应环境信号的稳态软骨细胞;以及未曾报道过的4种亚群:在退变髓核中富含的可能参与血管生成的髓核软骨细胞亚群C1;表达与细胞外基质分泌相关的基因亚群C2;仅在退变样本中检测到高表达基因参与炎症免疫反应和细胞外基质重塑的亚群C3;表达可维持胶原蛋白稳态的基因与成纤维细胞显示出相似表型的亚群C4等。作者推断软骨祖细胞的耗竭与C1亚群的转化是髓核退变的主要过程。 HAN等[35]对1个正常的髓核、3个定义为轻度退变和2个被诊断为重度退变的髓核进行了单细胞转录组的测序,同样发现随着椎间盘退变的发展,软骨祖细胞在髓核细胞中的比例显著下降;占据退变髓核组织中大部分而在正常髓核组织中占比较少的软骨细胞类型C1、C3,这种软骨细胞类型有正向调节脉管系统发育、抗菌体液反应和过渡金属离子稳态等功能,并被归类为炎症反应亚群;以及高表达Ⅱ型胶原蛋白和蛋白多糖,在所有髓核组织中共同存在的可以保持细胞外基质规则结构的细胞亚群C2,具有类似于成纤维细胞的表型、可能有助于细胞外基质重塑的细胞亚群C4。此外也发现了退变髓核组织中存在巨噬细胞与内皮细胞的浸润现象。进一步的转录调控分析也发现了在C1、C3中NF-κB家族显示出增强的调节活性;C4中有助于维持纤维特性的TCF4调节子活性更高;在细胞周期和DNA损伤修复中发挥重要作用的TFDP1、BRCA1和MYBL1是软骨祖细胞特异性调节子。 LI等[34]对1例正常的髓核组织和1例退变的髓核组织进行了研究,发现以下5种软骨细胞亚群:特征基因为COL2A1、COL9A2、COL11A2和CHRDL2的软骨细胞;特征基因为COL1A1、COL6A3、COL1A2、COL3A1和AQP1的纤维软骨细胞;特征基因为MGP、MT1G、GPX3、CDO1、MT2A和CLU的钙化抑制软骨细胞;特征基因为PTGES、TREM1、TIMP1、TIMP3的炎症软骨细胞;特征基因为FN1、SESN2、GDF15、COL15A1的钙化软骨细胞。这项研究发现人正常髓核组织中参与矿物质吸收、黏着斑和铁死亡的细胞通路在很大程度上被激活。相比之下,流体剪切应力、动脉粥样硬化相关通路、补体和凝血级联、细胞凋亡、黏着斑、PI3K-Akt 信号通路、矿物质吸收、HIF-1 信号通路、铁死亡和p53信号通路的激活程度在退变髓核组织中更为明显。其中,矿物质吸收、铁死亡、流体剪切应力、动脉粥样硬化相关通路、细胞凋亡和黏着斑可能是参与人髓核组织退变的重要过程。正常髓核组织中的细胞亚群拥有胶原纤维组织、凋亡过程的负调控和细胞黏附等功能,而退变组织中高比例的亚群有包括细胞外基质组织、骨骼系统发育、翻译起始和骨化等细胞功能。 GAO等[25]对小鼠髓核进行了测序,发现4个亚群:主要富含细胞黏附和转化生长因子β通路的亚群1;含有叶酸生物合成和PI3K-Akt通路的亚群2;包括柠檬酸循环、糖胺聚糖生物合成、硫代谢、不饱和脂肪酸的生物合成等代谢合成过程的亚群3;富集“抗炎基因”“糖酵解基因”“糖鞘脂生物合成”和“蛋白质消化和吸收”等功能的亚群4。这些数据表明,小鼠髓核细胞由异质亚群组成,并提供了对这些不同亚群的功能、基因表达水平、细胞外基质调节和代谢稳态的新见解。 TU等[24]对8例不同退变程度髓核的测序得到了以下功能的髓核亚群:标记基因为FRZB、DKK的肥大软骨细胞样髓核细胞;表达编码参与基因细胞代谢蛋白质的MSMO1、HMGCS1的效应髓核细胞;标记基因为RPS29、RPS21的稳态髓核细胞;标记基因为CHI3L1、CXCL2、NFKB的调节髓核细胞;表达编码与纤维化相关的COL1A1、COL3A1、COL6A1的纤维髓核细胞;表达与细胞黏附和迁移相关的FN1、CRTAC1的黏附髓核细胞。进一步的细胞功能富集分析发现了不同亚群的不同功能:效应髓核细胞富含甾醇生物合成和糖胺聚糖代谢过程相关基因和细胞外基质组装的正调节;稳态髓核细胞富含与细胞稳态相关的过程,包括翻译调节和蛋白质/RNA代谢;调节髓核细胞富含负责细胞对炎症和内源性刺激反应的高表达基因;纤维髓核细胞具有祖细胞特性。肥大软骨样髓核细胞是所有退变髓核组织中的主要细胞类型,调节髓核细胞在轻度退变髓核组织中大量出现,稳态髓核细胞在中度退变髓核中大量出现并且随着退变严重程度减少;而纤维和黏附髓核细胞在严重退变髓核组织中大量出现。 铁死亡是区别于细胞坏死、细胞凋亡、细胞自噬的新的细胞程序性死亡[48]。铁死亡的主要机制是二价铁或酯氧合酶催化细胞膜上高表达的不饱和脂肪酸,使其发生脂质过氧化,从而诱导细胞的死亡;此外,还表现为抗氧化体系(谷胱甘肽和谷胱甘肽过氧化物酶4)的表达量降低[49]。最近的研究表明铁死亡与神经退行性疾病、癌症、中风和脑缺血再灌注损伤有关。WELCH等[42]和LI等[44]对人退变髓核的单细胞测序研究均报道了铁死亡在特定退变髓核细胞亚群中出现了功能富集。ZHANG等[28]也注意到特定髓核组织亚群中出现了铁死亡的功能富集,并进一步在大鼠椎间盘退变动物模型中观察到退变髓核组织铁积累、血红素加氧酶1的高表达和轻链铁蛋白的表达降低,同时发现与铁死亡相关的基因在轻度椎间盘退变组织中显著富集,由此推断铁死亡可能在退变早期发挥作用。 目前对于髓核组织研究的共识为髓核组织中绝大部分的细胞为软骨细胞或者软骨样细胞,以及少量的内皮细胞和巨噬细胞。髓核细胞在单细胞图谱上无法区分出显著差异性,更深一步的分层可得到有功能富集上差异或者不同表型的亚群。不同的研究者对髓核组织细胞亚群的划分和命名各有不同,但仍可总结为以下几类:一是在正常髓核组织中占比较多的髓核细胞,通常被命名为“纤维髓核细胞”,其具有多分化能力以保持纤维软骨细胞发育;二是在轻度退变髓核中大量出现的“稳态髓核细胞”,此亚群细胞主要调节软骨细胞代谢活动的稳态平衡以响应环境信号,并且高表达胶原蛋白相关基因与细胞外基质的分泌有关;三是仅在退变髓核组织中发现的“炎症髓核细胞”,在此亚群中NF-κB家族转录因子调节活性增强;四是在严重退变髓核组织中较多标记基因为FN1的“黏附髓核细胞”。正常、退变髓核包含不同功能亚群的比例与类型各不相同。这些对髓核细胞异质性的分析揭示除了髓核组织本身的异质性,对于不同细胞的功能分析有助于更好地认知髓核退变的过程和机制。 2.2.3 髓核祖细胞 脊索细胞一直被认为是所有髓核细胞的前体[50],既往的研究认为脊索细胞在人类的青春期时开始消失[51],然而有研究发现成人髓核细胞中仍有脊索细胞的存在[52-53]。除此之外,间充质干细胞的标记基因CD44、NT5E(CD73)、THY1(CD90)、ENG(CD105)、NGFR(CD271)、NGFR(CD271)也在椎间盘中有表达[54]。之前的批量转录组测序研究无法揭示髓核组织内的异质性,使得寻找髓核祖细胞变得困难。单细胞测序技术的出现使得这一目的逐步达成。 GAN等[27]对人类健康髓核的测序发现了一组高表达PDGFRA、PRRX1和IGF1的髓核细胞亚群。其中,PDGFRA是间充质祖细胞的标志物;PPRX1是仅存在中胚层起源和间充质前体的调节物;IGF1是一种促进间充质干细胞分化为髓核样细胞的生长因子。因此将PDGFRA阳性的细胞亚群鉴定为髓核祖细胞。对这群细胞的进一步分析发现了同时表达PROCR和PDGFRA的髓核祖细胞。用流式细胞技术从原代髓核细胞中分选出PROCR+PDGFRA+髓核细胞进行集落形成单位-成纤维细胞集落形成测定,发现相比较PROCR-的髓核细胞,PROCR+PDGFRA+髓核细胞有显著增强的集落形成能力。 JIANG等[32]对新生儿和成人的椎间盘进行了转录组测序,发现新生儿和成人髓核中的脊索细胞有着不同的生物学特性。相比较髓核细胞,脊索细胞优先富集细胞增殖、细胞代谢、细胞稳态、血管生成以及细胞黏附和扩散,而髓核细胞优先富集细胞骨架组织形成、细胞质发育、创伤愈合和细胞外基质生成。新生儿髓核中的脊索细胞高表达SOX17和ITGA6,而成人的脊索细胞高表达CD44,并且鉴定出AKR1C1、APOE和FABP4作为人类椎间盘脊索细胞的新标志物。 GAO等[25]对小鼠髓核的测序发现了一组富集“正向调控细胞迁移”和“调控细胞生长”,对转化生长因子β、表皮生长因子等参与细胞分化、发育和稳态的多种信号通路具有高度响应性的髓核祖细胞亚群,并将尿紧张素Ⅱ(UTS2R)鉴定为其表面标记物。体外实验验证了UTS2R阳性髓核细胞具有良好的干性。UTS2R阳性髓核细胞在椎间盘退变过程中逐渐耗尽,这表明髓核祖细胞可能在维持生理椎间盘稳态和调节人类椎间盘退变进展中发挥重要作用。 TU等[24]对8例人退变髓核的单细胞测序研究发现一组可能具有祖细胞特性的CD90阳性髓核细胞。油红O染色和茜素红染色显示CD90阳性髓核细胞具有分化成多种细胞谱系包括成骨细胞和脂肪细胞的能力。与CD90阴性髓核细胞相比,CD90阳性髓核细胞显著表达纤维髓核细胞的表现基因。在促软骨分化的培养基中培养CD90阳性髓核细胞6 d后可分化为其他表型的髓核细胞。另外还发现从轻度退变髓核中分离出的CD90阳性髓核细胞比重度退变髓核中分离出的细胞具有更高的生长和分化潜力。这些表明CD90阳性髓核细胞可能是退变髓核中的祖细胞。 另外,WANG等[29]对大鼠椎间盘测序研究中根据追踪间充质干细胞定位出了一群表达Mcam、Eng、Tie等的细胞群,分离出来的细胞群可以在体外状态下进行骨与软骨的分化。 髓核细胞增殖缓慢,缺乏自我再生能力,增加了腰椎间盘退变治疗难度。目前的治疗手段着重于缓解椎间盘退变的临床症状,研究人员试图激活内源性髓核祖细胞或者移植具有活性外源性髓核祖细胞以修复椎间盘或者延缓退变。此类细胞在组织中的数量较少,应用以往的bulk转录组测序法无法寻找内源性髓核祖细胞,而单细胞转录组测序的出现使得研究人员顺利发现祖细胞的存在。通过单细胞转录组测序,以上研究利用间充质多功能干细胞的基因追踪以及细胞亚群的功能富集分析,寻找到不止一组具有强烈增殖、分化能力的髓核祖细胞,同时体外实验验证更体现出这些祖细胞成为修复或延缓椎间盘退变工程细胞的可能。在TU等[24]公布到公共数据库中的单细胞测序数据中发现:高表达CD90(THY1)的髓核细胞与PROCR+PDGFRA+髓核细胞在UMAP分布图上高度重叠,见图4,这说明两者可能发现了同一组髓核祖细胞。目前仍未有实验验证祖细胞在体内是否可以修复退变髓核组织。这些结论为髓核组细胞的存在提供大量证据,使得利用再生医学与组织工程作为髓核退变的治疗靶点成为可能。"

2.2.4 退变髓核的免疫细胞浸润 正常的椎间盘组织中缺乏巨噬细胞或中性粒细胞等免疫细胞[55]。而在一些动物实验和基础实验中发现退变椎间盘有免疫细胞的浸润[56]。单细胞测序使得研究者可以更直观地探究在组织中占少量细胞群的类型。ZHANG等[28]的研究首次报道了在单细胞测序维度下退变髓核的巨噬细胞浸润现象。紧接着,LI等[34]与HAN等[35]的研究也证明了人退变髓核中有免疫细胞浸润的现象。 WANG等[57]通过生物信息学分析鉴定了参与椎间盘组织髓核退变免疫浸润的关键基因,并推断出特定髓核亚群中表达的关键基因以及浸润性巨噬细胞在椎间盘退行性变的发病机制中起重要作用。 LING等[33]对髓核组织的免疫浸润进行了更为详细的研究:一项对3个不同退变程度的髓核进行单细胞测序的研究鉴定了包括巨噬细胞、T细胞、骨髓祖细胞和中性粒细胞在内的免疫细胞,巨噬细胞和髓核祖细胞在退变过程中通过巨噬细胞迁移抑制因子和NF-κB信号通路发生了显著的细胞间相互作用,并且巨噬细胞的极化可能在椎间盘退变放大炎症级联反应中起重要作用。 TU等[24]发现了在人退变髓核中除单核巨噬细胞、NK细胞、T细胞、粒细胞之外的一种粒-髓源性抑制细胞(granulocytic myeloid-derived suppressor cell,G-MDSC)。通过流式细胞技术,在体外和体内证明了G-MDSC在轻度退变髓核组织中的数量较重度退变髓核组织中更多,发现了相比较CD24- G-MDSC,CD24+ G-MDSC有着更为显著的T细胞抑制性和更强的活性氧产生能力;通过与髓核细胞共培养,发现其可以减缓白细胞介素1诱导的髓核组织退变。 在以上研究中发现,一些免疫细胞在退变椎间盘中的浸润增加。据报道,随着椎间盘退行性分级的进展,巨噬细胞的浸润明显加强。巨噬细胞、T细胞和中性粒细胞会释放肿瘤坏死因子α、白细胞介素1 a/b、白细胞介素6、白细胞介素17、白细胞介素8、白细胞介素2、白细胞介素4、白细胞介素10等细胞因子,促进椎间盘源性疼痛以及椎间盘退变。单细胞测序直接检测出免疫细胞在退变椎间盘中的浸润现象,进一步研究表明其与椎间盘源性疼痛相关[8]。越来越多的稀有细胞亚群及相关致病机制被发现,期待从免疫细胞浸润角度治疗椎间盘退行性变的研究出现。 2.2.5 纤维环的异质性 虽然大部分研究认为髓核的退变是椎间盘退变的主要原因,但仍有部分学者认为纤维环变性是椎间盘退行性变的关键,纤维环的变性直接导致髓核突出,进而导致炎症因子释放,压迫神经根和脊髓[58]。既往的研究表明,最外层的纤维环与靠近髓核的纤维环之间有异质性。CALIó等[31]对牛尾椎间盘的单细胞测序研究表明纤维环组织具有显著的异质性。 WANG等[26]对小鼠纤维化进行单细胞转录组测序的研究发现了10个主要的细胞亚群:纤维环干细胞、软骨样纤维环祖细胞、成纤维细胞样纤维环祖细胞、前软骨细胞样纤维环细胞、前成纤维细胞样纤维环细胞、未成熟软骨细胞样纤维环细胞、未成熟成纤维细胞样纤维环细胞、成熟软骨细胞样纤维环细胞、成熟成纤维细胞样纤维环细胞和纤维软骨细胞样纤维环细胞。使用免疫荧光验证了不同亚群的细胞在空间分布的区别,并且发现纤维环干细胞具有分化为纤维环细胞的潜力;接着使用β-甘油磷酸酯、转化生长因子β3、抗坏血酸和地塞米松组成的纤维软骨诱导补充剂在体外诱导纤维环干细胞成功分化为纤维软骨细胞样纤维环细胞;在小鼠的体内实验中证明了丝素蛋白、透明质酸和纤维软骨诱导补充剂的复合材料可有效促进纤维环的重建。 从纤维环探索椎间盘退行性变的治疗方案是区别于从髓核角度着手的全新方向。上述研究创新性地开发了一种用于纤维环重建的新型生物活性材料,推动了纤维环修复技术的发展。"

| [1] GBD 2019 DISEASES AND INJURIES COLLABORATORS. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396(10258):1204-1222. [2] KATZ JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88 Suppl 2:21-24. [3] ZAINA F, TOMKINS-LANE C, CARRAGEE E, et al. Surgical versus non-surgical treatment for lumbar spinal stenosis. Cochrane Database Syst Rev. 2016;2016(1):CD010264. [4] HUNTER CJ, MATYAS JR, DUNCAN NA. Cytomorphology of notochordal and chondrocytic cells from the nucleus pulposus: a species comparison. J Anat. 2004;205(5):357-362. [5] BAGNALL KM, HIGGINS SJ, SANDERS EJ. The contribution made by cells from a single somite to tissues within a body segment and assessment of their integration with similar cells from adjacent segments. Development. 1989;107(4):931-943. [6] GOLDSTEIN RS, KALCHEIM C. Determination of epithelial half-somites in skeletal morphogenesis. Development. 1992;116(2):441-445. [7] VERGROESEN PP, KINGMA I, EMANUEL KS, et al. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthritis Cartilage. 2015;23(7):1057-1070. [8] ITO K, CREEMERS L. Mechanisms of intervertebral disk degeneration/injury and pain: a review. Global Spine J. 2013;3(3):145-152. [9] SAKAI D, NAKAI T, MOCHIDA J, et al. Differential phenotype of intervertebral disc cells: microarray and immunohistochemical analysis of canine nucleus pulposus and anulus fibrosus. Spine (Phila Pa 1976). 2009;34(14):1448-1456. [10] LUECKEN MD, THEIS FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15(6):e8746. [11] KIM C, GAO R, SEI E, et al. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell. 2018;173(4): 879-893.e13. [12] HO DW, TSUI YM, CHAN LK, et al. Single-cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of HBV-associated hepatocellular carcinoma. Nat Commun. 2021;12(1):3684. [13] GUO J, GROW EJ, MLCOCHOVA H, et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28(12):1141-1157. [14] ZHONG S, ZHANG S, FAN X, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555(7697):524-528. [15] HAN X, CHEN H, HUANG D, et al. Mapping human pluripotent stem cell differentiation pathways using high throughput single-cell RNA-sequencing. Genome Biol. 2018;19(1):47. [16] HWANG B, LEE JH, BANG D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50(8):1-14. [17] BAGERITZ J, RADDI G. Single-Cell RNA Sequencing with Drop-Seq. Methods Mol Biol. 2019;1979:73-85. [18] FAN HC, FU GK, FODOR SP. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015; 347(6222):1258367. [19] DING J, ADICONIS X, SIMMONS SK, et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol. 2020;38(6):737-746. [20] LIANG X, SHEN H, SHI WD, et al. Effect of axial vertical vibration on degeneration of lumbar intervertebral discs in modified bipedal rats: An in-vivo study. Asian Pac J Trop Med. 2017;10(7):714-717. [21] TEKARI A, CHAN SCW, SAKAI D, et al. Angiopoietin-1 receptor Tie2 distinguishes multipotent differentiation capability in bovine coccygeal nucleus pulposus cells. Stem Cell Res Ther. 2016;7(1):75. [22] CHE YJ, GUO JB, LIANG T, et al. Assessment of changes in the micro-nano environment of intervertebral disc degeneration based on Pfirrmann grade. Spine J. 2019;19(7):1242-1253. [23] GRIFFITH JF, WANG YX, ANTONIO GE, et al. Modified Pfirrmann grading system for lumbar intervertebral disc degeneration. Spine (Phila Pa 1976). 2007;32(24):E708-712. [24] TU J, LI W, YANG S, et al. Single-Cell Transcriptome Profiling Reveals Multicellular Ecosystem of Nucleus Pulposus during Degeneration Progression. Adv Sci (Weinh). 2022;9(3):e2103631. [25] GAO B, JIANG B, XING W, et al. Discovery and application of postnatal nucleus pulposus progenitors essential for intervertebral disc homeostasis and degeneration. Adv Sci (Weinh). 2022;9(13):e2104888. [26] WANG H, WANG D, LUO B, et al. Decoding the annulus fibrosus cell atlas by scrna-seq to develop an inducible composite hydrogel: a novel strategy for disc reconstruction. Bioact Mater. 2022;14:350-363. [27] GAN Y, HE J, ZHU J, et al. Spatially defined single-cell transcriptional profiling characterizes diverse chondrocyte subtypes and nucleus pulposus progenitors in human intervertebral discs. Bone Res. 2021; 9(1):37. [28] ZHANG Y, HAN S, KONG M, et al. Single-cell RNA-seq analysis identifies unique chondrocyte subsets and reveals involvement of ferroptosis in human intervertebral disc degeneration. Osteoarthritis Cartilage. 2021;29(9):1324-1334. [29] WANG J, HUANG Y, HUANG L, et al. Novel biomarkers of intervertebral disc cells and evidence of stem cells in the intervertebral disc. Osteoarthritis Cartilage. 2021;29(3):389-401. [30] CHERIF H, MANNARINO M, PACIS AS, et al. Single-Cell RNA-Seq Analysis of Cells from Degenerating and Non-Degenerating Intervertebral Discs from the Same Individual Reveals New Biomarkers for Intervertebral Disc Degeneration. Int J Mol Sci. 2022;23(7):3993. [31] CALIÓ M, GANTENBEIN B, EGLI M, et al. The Cellular Composition of Bovine Coccygeal Intervertebral Discs: A Comprehensive Single-Cell RNAseq Analysis. Int J Mol Sci. 2021;22(9):4917. [32] JIANG W, GLAESER JD, SALEHI K, et al. Single-cell atlas unveils cellular heterogeneity and novel markers in human neonatal and adult intervertebral discs. iScience. 2022;25(7):104504. [33] LING Z, LIU Y, WANG Z, et al. Single-Cell RNA-Seq Analysis Reveals Macrophage Involved in the Progression of Human Intervertebral Disc Degeneration. Front Cell Dev Biol. 2022;9:833420. [34] LI Z, YE D, DAI L, et al. Single-Cell RNA Sequencing Reveals the Difference in Human Normal and Degenerative Nucleus Pulposus Tissue Profiles and Cellular Interactions. Front Cell Dev Biol. 2022;10:910626. [35] HAN S, ZHANG Y, ZHANG X, et al. Single-cell rna sequencing of the nucleus pulposus reveals chondrocyte differentiation and regulation in intervertebral disc degeneration. Front Cell Dev Biol. 2022;10:824771. [36] PANEBIANCO CJ, DAVE A, CHARYTONOWICZ D, et al. Single-cell RNA-sequencing atlas of bovine caudal intervertebral discs: Discovery of heterogeneous cell populations with distinct roles in homeostasis. FASEB J. 2021;35(11):e21919. [37] MANGIOLA S, DOYLE MA, PAPENFUSS AT. Interfacing Seurat with the R tidy universe. Bioinformatics. 2021;37(22):4100-4107. [38] MCCARTHY DJ, CAMPBELL KR, LUN AT, et al. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33(8):1179-1186. [39] WOLF FA, ANGERER P, THEIS FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. [40] DAL MOLIN A, DI CAMILLO B. How to design a single-cell RNA-sequencing experiment: pitfalls, challenges and perspectives. Brief Bioinform. 2019;20(4):1384-1394. [41] KORSUNSKY I, MILLARD N, FAN J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12): 1289-1296. [42] WELCH JD, KOZAREVA V, FERREIRA A, et al. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell. 2019;177(7):1873-1887.e17. [43] HAGHVERDI L, LUN ATL, MORGAN MD, et al. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36(5):421-427. [44] LI X, WANG K, LYU Y, et al. Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat Commun. 2020;11(1):2338. [45] CHEN W, ZHAO Y, CHEN X, et al. A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples. Nat Biotechnol. 2021;39(9):1103-1114. [46] TRAN HTN, ANG KS, CHEVRIER M, et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21(1):12. [47] RIESTER SM, LIN Y, WANG W, et al. RNA sequencing identifies gene regulatory networks controlling extracellular matrix synthesis in intervertebral disk tissues. J Orthop Res. 2018;36(5):1356-1369. [48] DIXON SJ, LEMBERG KM, LAMPRECHT MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060-1072. [49] TANG D, KANG R, BERGHE TV, et al. The molecular machinery of regulated cell death. Cell Res. 2019;29(5):347-364. [50] RISBUD MV, SCHAER TP, SHAPIRO IM. Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev Dyn. 2010;239(8):2141-2148. [51] PAZZAGLIA UE, SALISBURY JR, BYERS PD. Development and involution of the notochord in the human spine. J R Soc Med. 1989;82(7):413-415. [52] RICHARDSON SM, LUDWINSKI FE, GNANALINGHAM KK, et al. Notochordal and nucleus pulposus marker expression is maintained by sub-populations of adult human nucleus pulposus cells through aging and degeneration. Sci Rep. 2017;7(1):1501. [53] LIU Z, ZHENG Z, QI J, et al. CD24 identifies nucleus pulposus progenitors/ notochordal cells for disc regeneration. J Biol Eng. 2018;12:35. [54] ERWIN WM, ISLAM D, EFTEKARPOUR E, et al. Intervertebral disc-derived stem cells: implications for regenerative medicine and neural repair. Spine (Phila Pa 1976). 2013;38(3):211-216. [55] LIN Y, CONG H, LIU K, et al. Microbicidal Phagocytosis of Nucleus Pulposus Cells Against Staphylococcus aureus via the TLR2/MAPKs Signaling Pathway. Front Immunol. 2019;10:1132. [56] SUN Z, LIU B, LUO ZJ. The immune privilege of the intervertebral disc: implications for intervertebral disc degeneration treatment. Int J Med Sci. 2020;17(5):685-692. [57] WANG L, HE T, LIU J, et al. Revealing the Immune Infiltration Landscape and Identifying Diagnostic Biomarkers for Lumbar Disc Herniation. Front Immunol. 2021;12:666355. [58] CHU G, SHI C, LIN J, et al. Biomechanics in Annulus Fibrosus Degeneration and Regeneration. Adv Exp Med Biol. 2018;1078:409-420. |
| [1] | Nong Fuxiang, Jiang Zhixiong, Li Yinghao, Xu Wencong, Shi Zhilan, Luo Hui, Zhang Qinglang, Zhong Shuang, Tang Meiwen. Bone cement augmented proximal femoral nail antirotation for type A3.3 intertrochanteric femoral fracturalysis [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(在线): 1-10. |
| [2] | He Xi, Wan Yu, Tang Yuting, Yang Anning, Wu Kai, Jiao Yun, Bai Zhigang, Jiang Yideng, Shen Jiangyong. Erastin inhibits proliferation of hypertrophic scar fibroblasts [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(在线): 1-. |
| [3] | Cao Sheng, Kong Lingwei, Xu Kun, Sun Zhijie. Correlation of cervical sagittal force line parameters with degenerative segment and Pfirrmann classification in patients with cervical intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(9): 1319-1324. |
| [4] | You Zhengqiu, Zhang Zhongzu, Wang Qunbo. Early symptomatic intervertebral disc pseudocysts after discectomy detected on MRI [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(9): 1403-1409. |
| [5] | Lian Shilin, Zhang Yan, Jiang Qiang, Zhang Hanshuo, Li Tusheng, Ding Yu. Interventional effects of whole blood and platelet-rich plasma with different preparation methods on nucleus pulposus cells [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(8): 1199-1204. |
| [6] | Xiong Zhilin, Sun Hong, Liu Miao, Zhuang Yong. Roles of ferroptosis in intervertebral disc degeneration and osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(36): 5884-5890. |
| [7] | Liang Xuezhen, Luo Di, Li Jiacheng, Wen Mingtao, Zhang Jian, Xu Bo, Li Gang. PTGS2 and STAT3 in steroid-induced osteonecrosis of the femoral head: ferroptosis-related potential diagnostic biomarkers [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(36): 5898-5904. |
| [8] | Zheng Xiaohan, Feng Xiaoli, Hu Lan, Gao Shijun, Wei Yanzhao, Huang Ting, Sun Shengtong, Wei Xufang, Wang Tan, Zhao Zhenqiang. Macrophage migration inhibitory factor promotes the differentiation of human embryonic stem cells into ventral midbrain dopaminergic neural progenitor cells [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(33): 5348-5356. |
| [9] | Zhang Shijing, Huang Zhuojian, Tao Quyuan, Hou Jiangtao, Peng Bin, Luo Yongxin, Li Yanwu, Li Huibiao, Zhong Jiamin, Chen Xinlin. Molecular characteristic of immune cells in ulcerative colitis patients with spleen deficiency and dampness syndrome based on single-cell RNA sequencing [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(33): 5357-5362. |
| [10] | He Xi, Wan Yu, Tang Yuting, Yang Anning, Wu Kai, Jiao Yun, Bai Zhigang, Jiang Yideng, Shen Jiangyong. Erastin inhibits proliferation of hypertrophic scar fibroblasts [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(32): 5120-5125. |
| [11] | Dong Jiaan, Liu Ruyin, Yue Zongjin, Xu Xiangyang, Wang Zhengzhen. Huangqi Guizhi Wuwu Decoction attenuates mitochondrial damage and oxidative stress in intervertebral disc endplate chondrocytes [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(32): 5137-5143. |
| [12] | Yang Chaoqiang, Zhang Hulin, Wang Xiaomin, Wang Liang, Wang Yican. Prevention and treatment of bone-related diseases by regulating macrophages with Chinese medicine [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(31): 5064-5070. |
| [13] | Liu Xin, Sun Tianze, Zhang Jing, Zhang Wentao, Li Zhonghai. Research and advance in intervertebral disc annulus fibrosus repair [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(31): 5078-5084. |
| [14] | Lu Xiaojun, Xiong Bohan, Yang Tengyun, Wang Xu, Zhang Yaozhang, Zhong Ruiying, Li Yanlin. Novel programmed cell death of chondrocytes in osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(28): 4571-4576. |
| [15] | Xu Guangming, Liang Ziyang, Wang Hongbo, Zhang Zhen, Xiao Qinghua, Yang Jiyong, Lin Xiaosheng. Mechanical effect of oblique-pulling manipulation on triarticular complex after human lumbar intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(27): 4277-4282. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||