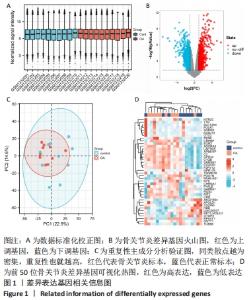
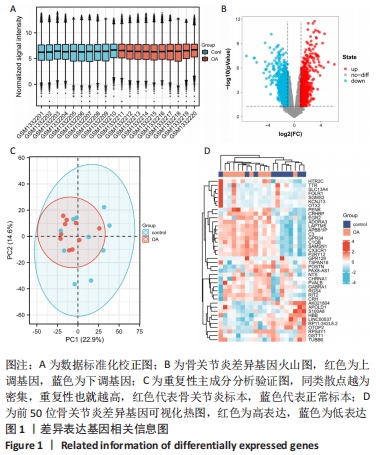
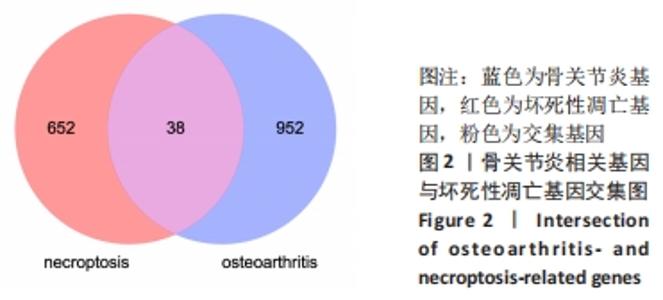
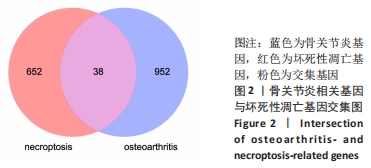
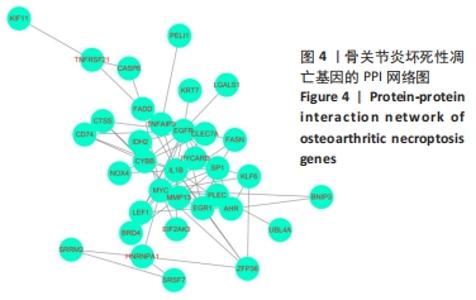
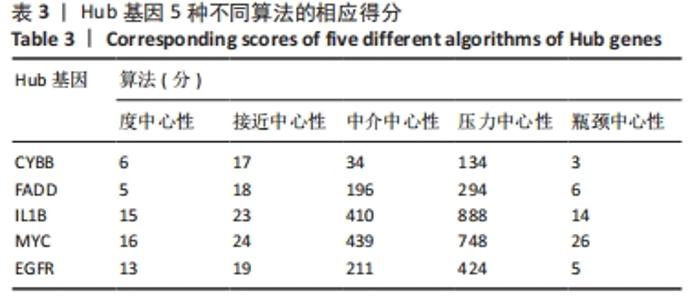
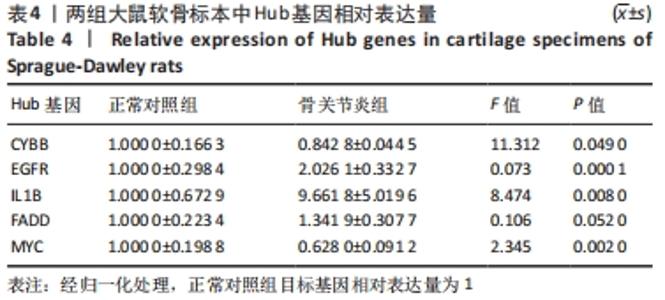
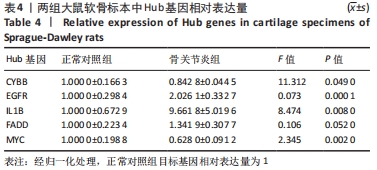
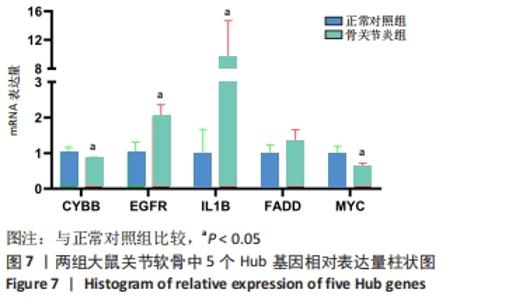
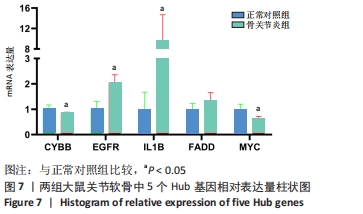
[1] HUNTER DJ, BIERMA-ZEINSTRA S. Osteoarthritis. Lancet. 2019;393(10182):1745-1759.
[2] NELSON AE. Osteoarthritis year in review 2017: clinical. Osteoarthritis Cartilage. 2018;26(3):319-325.
[3] SHEN J, ABU-AMER Y, O’KEEFE RJ, et al. Inflammation and epigenetic regulation in osteoarthritis. Connect Tissue Res. 2017;58(1):49-63.
[4] LATOURTE A, KLOPPENBURG M, RICHETTE P. Emerging pharmaceutical therapies for osteoarthritis. Nat Rev Rheumatol. 2020;16(12):673-688.
[5] 董方,商建峰,方微,等.坏死性凋亡在心血管疾病发生发展中的作用及其机制进展[J].中华心血管病杂志,2021,49(7):728-732.
[6] CHENG J, DUAN X, FU X, et al. RIP1 Perturbation Induces Chondrocyte Necroptosis and Promotes Osteoarthritis Pathogenesis via Targeting BMP7. Front Cell Dev Biol. 2021;9:638382.
[7] RIEGGER J, BRENNER RE. Evidence of necroptosis in osteoarthritic disease: investigation of blunt mechanical impact as possible trigger in regulated necrosis. Cell Death Dis. 2019;10(10):1-12.
[8] JEON J, NOH HJ, LEE H, et al. TRIM24-RIP3 axis perturbation accelerates osteoarthritis pathogenesis. Ann Rheum Dis. 2020;79(12):1635-1643.
[9] WANG J, CHEN L, JIN S, et al. MiR-98 promotes chondrocyte apoptosis by decreasing Bcl-2 expression in a rat model of osteoarthritis. Acta Biochim Biophys Sin (Shanghai). 2016;48(10):923-929.
[10] 王云峰,白人骁,张扬,等.改良Hulth模型复制膝不同时期骨关节炎的实验研究[J].天津医科大学学报,2009,15(3):400-404.
[11] LEE SW, RHO JH, LEE SY, et al. Leptin protects rat articular chondrocytes from cytotoxicity induced by TNF-α in the presence of cyclohexamide. Osteoarthritis Cartilage. 2015;23(12):2269-2278.
[12] YANG T, CAO C, YANG J, et al. miR-200a-5p regulates myocardial necroptosis induced by Se deficiency via targeting RNF11. Redox Biol. 2018;15:159-169.
[13] LI X, YAO X, ZHU Y, et al. The Caspase Inhibitor Z-VAD-FMK Alleviates Endotoxic Shock via Inducing Macrophages Necroptosis and Promoting MDSCs-Mediated Inhibition of Macrophages Activation. Front Immunol. 2019;10:1824.
[14] KOURTZELIS I, MITROULIS I, VON RENESSE J, et al. From leukocyte recruitment to resolution of inflammation: the cardinal role of integrins. Leukoc Biol. 2017; 102(3):677-683.
[15] JAYARAM P, KANG GE, HELDT BL, et al. Novel assessment of leukocyte-rich platelet-rich plasma on functional and patient-reported outcomes in knee osteoarthritis: a pilot study. Regen Med. 2021;16(9):823-832.
[16] GUO N, YE S, ZHANG K, et al. A critical epitope in CD147 facilitates memory CD4+ T-cell hyper-activation in rheumatoid arthritis. Cell Mol Immunol. 2019; 16(6):568-579.
[17] ROSSHIRT N, TRAUTH R, PLATZER H, et al. Proinflammatory T cell polarization is already present in patients with early knee osteoarthritis. Arthritis Res Ther. 2021;23(1):37.
[18] TAKEUCHI Y, OHARA D, WATANABE H, et al. Dispensable roles of Gsdmd and Ripk3 in sustaining IL-1β production and chronic inflammation in Th17-mediated autoimmune arthritis. Sci Rep. 2021;11(1):18679.
[19] NA HS, PARK JS, CHO KH, et al. Interleukin-1-Interleukin-17 Signaling Axis Induces Cartilage Destruction and Promotes Experimental osteoarthritis. Front Immunol. 2020;11:730.
[20] GUTIERREZ KD, DAVIS MA, DANIELS BP, et al. MLKL Activation Triggers NLRP3-Mediated Processing and Release of IL-1β Independently of Gasdermin-D. J Immunol. 2017;198(5):2156-2164.
[21] LIAO CR, WANG SN, ZHU SY, et al. Advanced oxidation protein products increase TNF-α and IL-1β expression in chondrocytes via NADPH oxidase 4 and accelerate cartilage degeneration in osteoarthritis progression. Redox Biol. 2020;28:101306.
[22] ZHOU S, ZHANG W, CAI G, et al. Myofiber necroptosis promotes muscle stem cell proliferation via releasing Tenascin-C during regeneration. Cell Res. 2020;30(12): 1063-1077.
[23] JIANG L, ZHOU X, XU K, et al. miR-7/EGFR/MEGF9 axis regulates cartilage degradation in osteoarthritis via PI3K/AKT/mTOR signaling pathway. Bioengineered. 2021;12(1):8622-8634.
[24] SEONG D, JEONG M, SEO J, et al. Identification of MYC as an antinecroptotic protein that stifles RIPK1-RIPK3 complex formation. Proc Natl Acad Sci U S A. 2020;117(33):19982-19993.
[25] JIANG K, JIANG T, CHEN Y, et al. Mesenchymal Stem Cell-Derived Exosomes Modulate Chondrocyte Glutamine Metabolism to Alleviate Osteoarthritis Progression. Mediators Inflamm. 2021;2021:2979124.
[26] LICHÝ M, SZOBI A, HRDLIČKA J, et al. Different signalling in infarcted and non-infarcted areas of rat failing hearts: A role of necroptosis and inflammation. Cell Mol Med. 2019;23(9):6429-6441.
[27] COLLINS JA, WOOD ST, NELSON KJ, et al. Oxidative Stress Promotes Peroxiredoxin Hyperoxidation and Attenuates Pro-survival Signaling in Aging Chondrocytes. J Biol Chem. 2016;291(13):6641-6654.
[28] DERAGON MA, MCCAIG WD, PATEL PS, et al. Mitochondrial ROS prime the hyperglycemic shift from apoptosis to necroptosis. Cell Death Discov. 2020;6(1): 132.
|