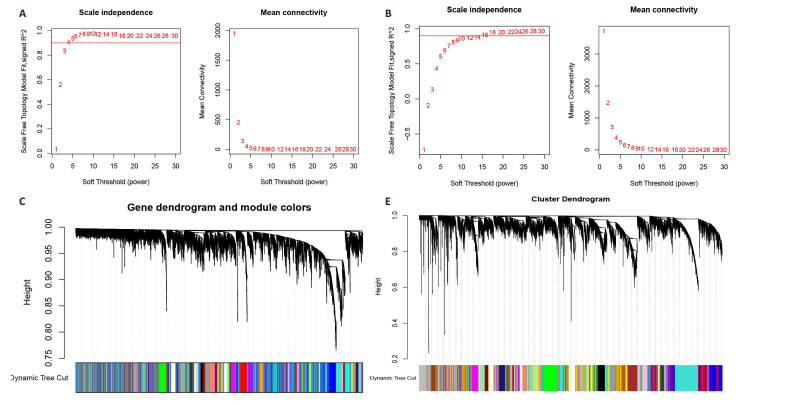
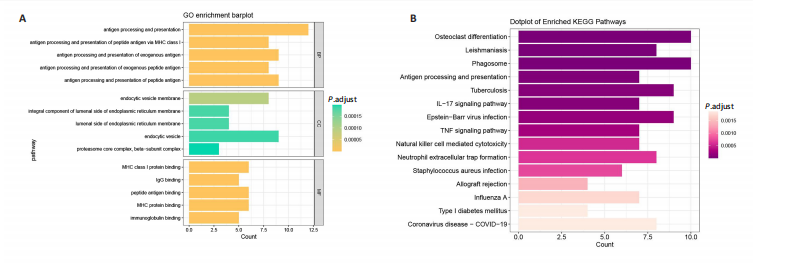
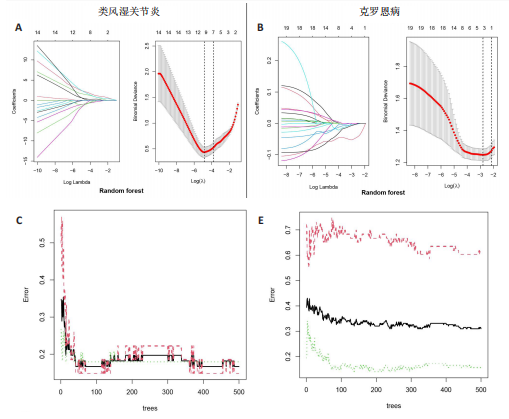
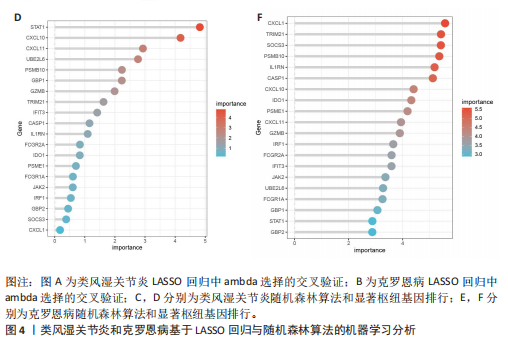
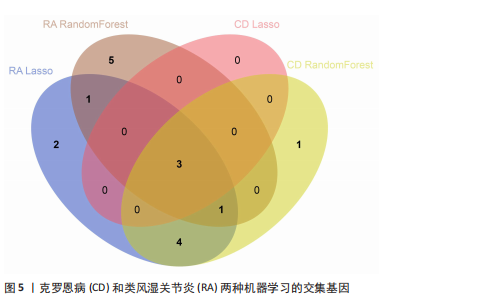
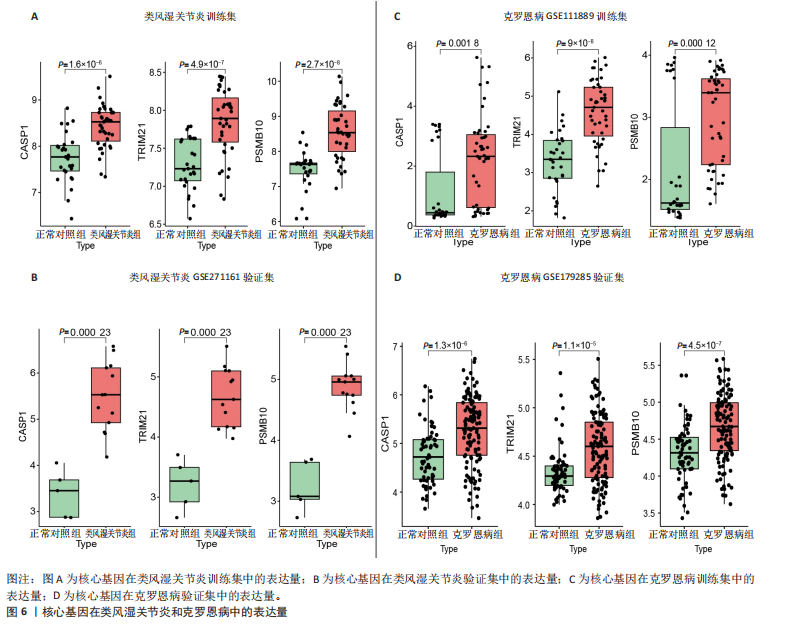
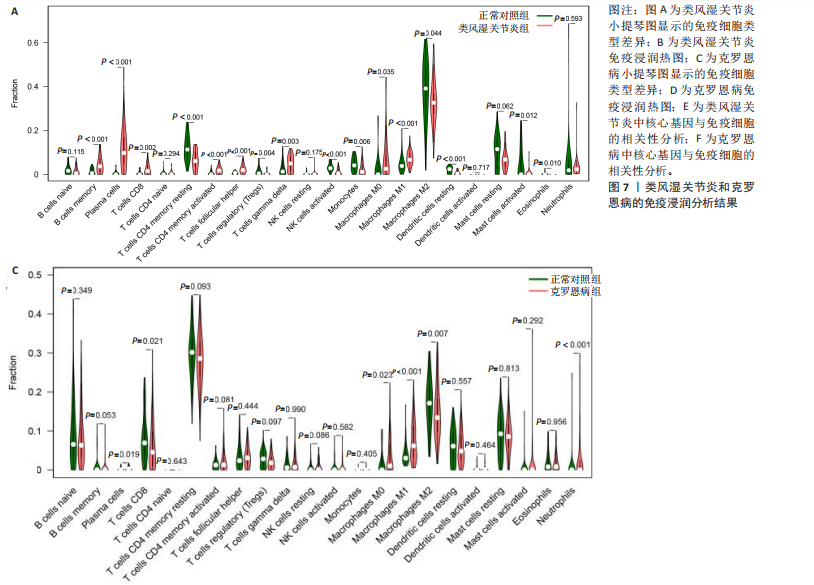
[1] CONFORTI A, DI COLA I, PAVLYCH V, et al. Beyond the joints, the extra-articular manifestations in rheumatoid arthritis. Autoimmun Rev. 2021;20(2):102735.
[2] TURESSON C, MATTESON EL. Extra-articular manifestations in rheumatoid arthritis. Int J Adv Rheumatol. 2007;5(3):72.
[3] RADU AF, BUNGAU SG. Management of rheumatoid arthritis: an overview. Cells. 2021;10(11):2857.
[4] WU Z, MA D, YANG H,et al. Fibroblast-like synoviocytes in rheumatoid arthritis: surface markers and phenotypes. Int Immunopharmacol. 2021;93:107392.
[5] RODA G, CHIEN NG S, KOTZE PG, et al. Crohn’s disease. Nat Rev Dis Primers. 2020; 6(1):22.
[6] PETAGNA L, ANTONELLI A, GANINI C, et al. Pathophysiology of Crohn’s disease inflammation and recurrence. Biol Direct. 2020;15(1):23.
[7] CLAYTOR J, KUMAR P, ANANTHAKRISHNAN AN, et al. Mild Crohn’s disease: definition and management. Curr Gastroenterol Rep. 2023;25(3):45-51.
[8] WANG B, XIONG Y, LI R, et al. Potential role of SNP rs2071475 in rheumatoid arthritis and inflammatory bowel disease in the East Asian population: a Mendelian randomization study. Inflammopharmacology. 2024;32(1):683-692.
[9] SACCON TD, DHAHBI JM, SCHNEIDER A, et al. Plasma miRNA profile of Crohn’s disease and rheumatoid arthritis patients. Biology. 2022;11(4):508.
[10] ELSOURI K, ARBOLEDA V, HEISER S, et al. Microbiome in rheumatoid arthritis and celiac disease: a friend or foe. Cureus. 2021;13(6):le15543.
[11] SCHER JU, ABRAMSON SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7(10):569-578.
[12] WANG Q, ZHANG SX, CHANG MJ,et al. Characteristics of the gut microbiome and its relationship with peripheral CD4+ T cell subpopulations and cytokines in rheumatoid arthritis. Frontiers in Microbiology. 2022; 13:799602.
[13] WELLS PM, WILLIAMS FM, MATEY-HERNANDEZ ML, et al.’RA and the microbiome: do host genetic factors provide the link? J Autoimmun. 2019;99:104-115.
[14] MASSIMINO L, BARCHI A, MANDARINO FV, et al. A multi-omic analysis reveals the esophageal dysbiosis as the predominant trait of eosinophilic esophagitis. J Transl Med. 2023;21(1):46.
[15] GURTNER A, CREPAZ D, ARNOLD IC. Emerging functions of tissue-resident eosinophils. J Exp Med. 2023;220(7): e20221435.
[16] JACOBS I, CEULEMANS M, WAUTERS L, et al. Role of eosinophils in intestinal inflammation and fibrosis in inflammatory bowel disease: an overlooked villain? Front Immunol. 2021;12:754413.
[17] FILIPPONE RT, SAHAKIAN L, APOSTOLOPOULOS V, et al. Eosinophils in inflammatory bowel disease. Inflamm Bowel Dis. 2019;25(7):1140-1151.
[18] ZOU C, BEARD JA, YANG G, et al. CASP orter: A Novel Inducible Human CASP1/NALP3/ASC Inflammasome Biosensor. J Inflamm Res. 2022;1:1183-1194.
[19] INSERRA A, CHOO JM, LEWIS MD, et al. Mice lacking Casp1, Ifngr and Nos2 genes exhibit altered depressive-and anxiety-like behaviour, and gut microbiome composition. Sci Rep. 2019;9(1):6456.
[20] ADDOBBATI C, DA CRUZ HL, ADELINO JE, et al. Polymorphisms and expression of inflammasome genes are associated with the development and severity of rheumatoid arthritis in Brazilian patients. Inflamm Res. 2018;67:255-264.
[21] FOSS S, BOTTERMANN M, JONSSON A, et al.TRIM21—from intracellular immunity to therapy. Front Immunol. 2019;10:2049.
[22] SAFONOVA TN, ZAITSEVA GV, LOGINOV VI, et al. Association of polymorphisms of the TRIM21 gene with the severity of dry keratoconjunctivitis in rheumatoid arthritis and Sjogren’s disease. Vestn Oftalmol. 2019;135(5.Vyp.2):192-198.
[23] LEE AY, REED JH, GORDON TP. Anti-Ro60 and anti-Ro52/TRIM21: Two distinct autoantibodies in systemic autoimmune diseases. J Autoimmun. 2021;124:102724.
[24] LIU J, ZHANG C, XU D, et al. The ubiquitin ligase TRIM21 regulates mutant p53 accumulation and gain of function in cancer. J Clin Invest. 2023;133(6):e164354.
[25] LIU YX, WAN S, YANG XQ, et al. TRIM21 is a druggable target for the treatment of metastatic colorectal cancer through ubiquitination and activation of MST2. Cell Chem Biol. 2023;30(7):709-725.
[26] SARRABAY G, MÉCHIN D, SALHI A, et al. PSMB10, the last immunoproteasome gene missing for PRAAS. J Allergy Clin Immunol. 2020;145(3):1015-1017.
[27] XiAO X, FENG X, YOO JH, et al. The β-Grasp Domain of Proteasomal ATPase Mpa Makes Critical Contacts with the Mycobacterium tuberculosis 20S Core Particle to Facilitate Degradation. Msphere. 2022;7(5):e0027422.
[28] SHI CX, ZHU YX, BRUINS LA, et al. Proteasome subunits differentially control myeloma cell viability and proteasome inhibitor sensitivity. Mol Cancer Res. 2020; 18(10):1453-1464.
[29] GUIMARÃES G, GOMES MT, CAMPOS PC, et al. Immunoproteasome subunits are required for CD8+ T cell function and host resistance to Brucella abortus infection in mice. Infect Immun. 2018;86(3):10-128.
[30] WEHR P, PURVIS H, LAW SC, et al. Dendritic cells, T cells and their interaction in rheumatoid arthritis. Clin Exp Immunol. 2019;196(1):12-27.
[31] HE J, LI Y, CHEN J, et al. The relationships of CD8+ T cell subsets in RA patients with disease activity and clinical parameters. Int Immunopharmacol. 2023;114:109399.
[32] JONSSON AH, ZHANG F, DUNLAP G, et al.Granzyme K+ CD8 T cells form a core population in inflamed human tissue. Sci Transl Med. 2022;14(649):eabo0686.
[33] HUANG B, CHEN Z, GENG L, et al. Mucosal profiling of pediatric-onset colitis and IBD reveals common pathogenics and therapeutic pathways. Cell. 2019;179(5): 1160-1176.
[34] CASALEGNO GARDUÑO R, DÄBRITZ J. New insights on CD8+ T cells in inflammatory bowel disease and therapeutic approaches. Front Immunol. 2021;12:738762.
[35] LI CH, MA ZZ, JIAN LL, et al. Iguratimod inhibits osteoclastogenesis by modulating the RANKL and TNF-α signaling pathways. Int Immunopharmacol. 2021;90:107219.
[36] CAI L, MU YR, LIU MM, et al. Penta-acetyl Geniposide suppresses migration, invasion, and inflammation of TNF-α-stimulated rheumatoid arthritis fibroblast-like synoviocytes involving wnt/β-catenin signaling pathway. Inflammation. 2021; 44(6):2232-2245.
[37] SCHMITT H, NEURATH MF, ATREYA R. Role of the IL23/IL17 Pathway in Crohn’s Disease. Front Immunol. 2021;12:622934.
[38] LOH W, VERMEREN S. Anti-inflammatory neutrophil functions in the resolution of inflammation and tissue repair. Cells. 2022;11(24):4076.
[39] OLIVEIRA SR, DE ARRUDA JA, SCHNEIDER AH, et al. Neutrophil extracellular traps in rheumatoid arthritis and periodontitis: Contribution of PADI4 gene polymorphisms. J Clin Periodontol. 2024;51(4):452-463.
[40] WRIGHT HL, LYON M, CHAPMAN EA, et al.Rheumatoid arthritis synovial fluid neutrophils drive inflammation through production of chemokines, reactive oxygen species, and neutrophil extracellular traps. Front Immunol. 2021;11:584116.
[41] GARRIDO-TRIGO A, CORRALIZA AM, VENY M, et al. Macrophage and neutrophil heterogeneity at single-cell spatial resolution in human inflammatory bowel disease. Nat Commun. 2023;14(1):4506.
[42] CHEN H, WU X, XU C, et al. Dichotomous roles of neutrophils in modulating pathogenic and repair processes of inflammatory bowel diseases. Precis Clin Med. 2021;4(4):246-257.
[43] WÉRA O, LANCELLOTTI P, OURY C. The dual role of neutrophils in inflammatory bowel diseases. J Clin Med. 2016;5(12):118.
[44] SOUFLI I, TOUMI R, RAFA H, et al. Overview of cytokines and nitric oxide involvement in immuno-pathogenesis of inflammatory bowel diseases. World J Gastrointest Pharmacol Ther. 2016;7(3):353.
|