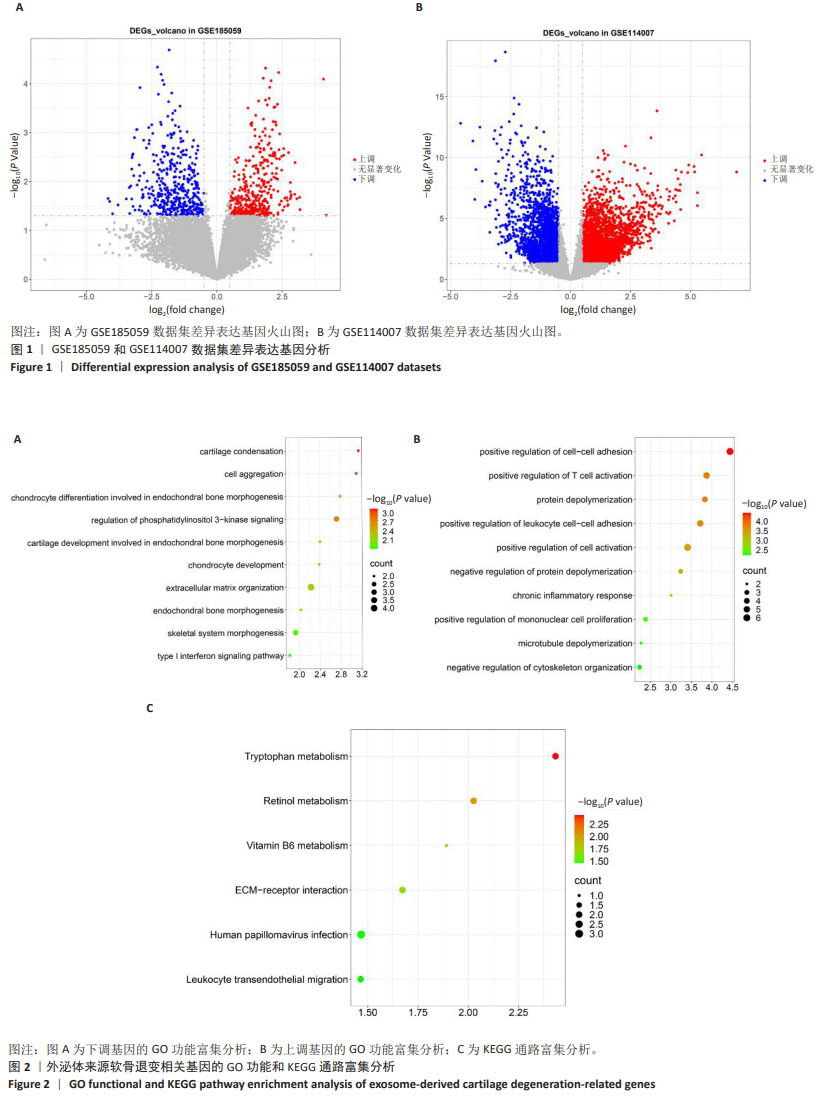
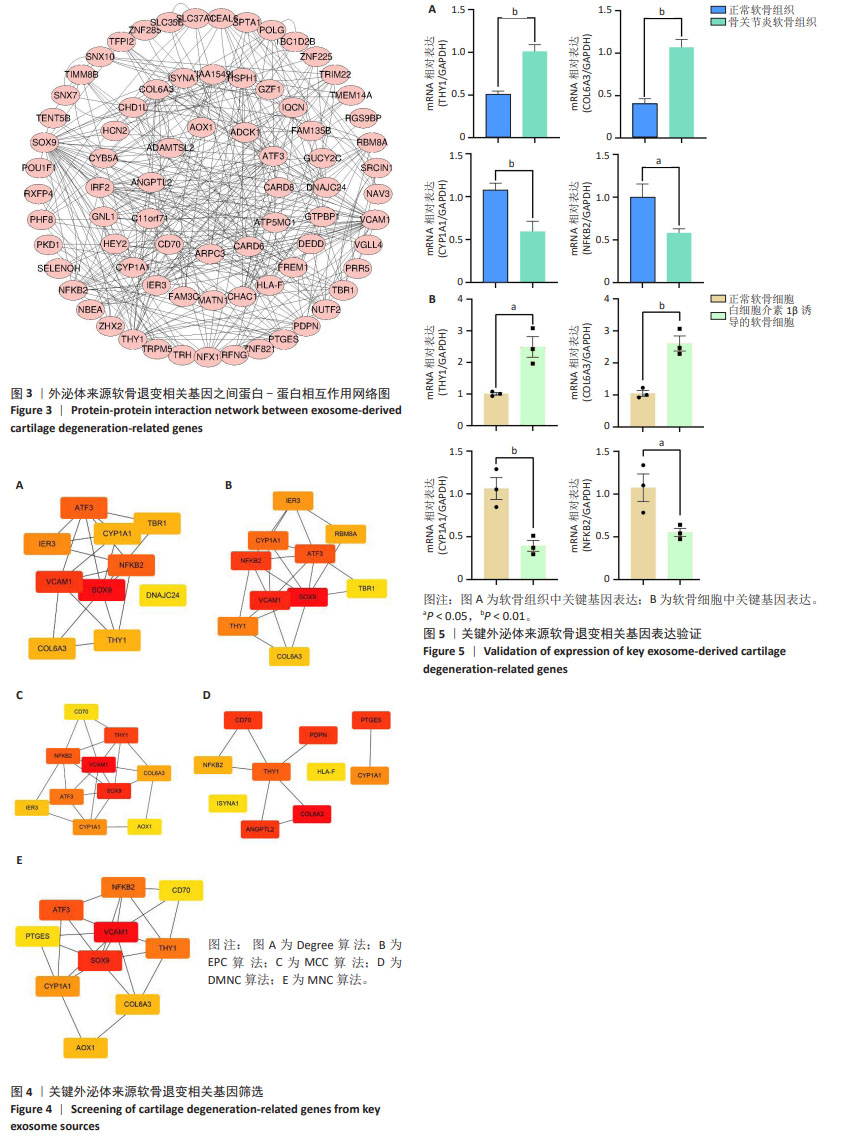
[1] MINNIG MCC, GOLIGHTLY YM, NELSON AE. Epidemiology of osteoarthritis: literature update 2022-2023. Curr Opin Rheumatol. 2024;36(2):108-112.
[2] 张烽,王斌,曹蔼萱,等.原发性膝关节骨关节炎严重程度的危险因素分析[J].中华创伤骨科杂志,2024,26(8):698-704.
[3] FUJII Y, LIU L, YAGASAKI L, et al. Cartilage Homeostasis and Osteoarthritis. Int J Mol Sci. 2022;23(11):6316.
[4] 章晓云,曾浩,孟林.膝骨关节炎疼痛机制及治疗研究进展[J].中国疼痛医学杂志,2023,29(1):50-58.
[5] 张琪,于湄,刘磊,等.工程化外泌体研究现状与临床转化的挑战[J].中国组织工程研究,2023,27(19):3052-3060.
[6] HE C, ZHENG S, LUO Y, et al. Exosome Theranostics: Biology and Translational Medicine. Theranostics. 2018;8(1):237-255.
[7] NI Z, KUANG L, CHEN H, et al. The exosome-like vesicles from osteoarthritic chondrocyte enhanced mature IL-1β production of macrophages and aggravated synovitis in osteoarthritis. Cell Death Dis. 2019;10(7):522.
[8] LI Z, WANG Y, XIANG S, et al. Chondrocytes-derived exosomal miR-8485 regulated the Wnt/β-catenin pathways to promote chondrogenic differentiation of BMSCs. Biochem Biophys Res Commun. 2020;523(2):506-513.
[9] JI Y, XIONG L, ZHANG G, et al. Synovial fluid exosome-derived miR-182-5p alleviates osteoarthritis by downregulating TNFAIP8 and promoting autophagy through LC3 signaling. Int Immunopharmacol. 2023; 125(Pt A):111177.
[10] LAI C, LIAO B, PENG S, et al. Synovial fibroblast-miR-214-3p-derived exosomes inhibit inflammation and degeneration of cartilage tissues of osteoarthritis rats. Mol Cell Biochem. 2023;478(3):637-649.
[11] 中华医学会骨科学分会关节外科学组.骨关节炎诊疗指南(2018年版)[J].中华骨科杂志,2018,38(12):705-715.
[12] LIU Q, HAN M, WU Z, et al. DDX5 inhibits hyaline cartilage fibrosis and degradation in osteoarthritis via alternative splicing and G-quadruplex unwinding. Nat Aging. 2024;4(5):664-680.
[13] MCCLURG O, TINSON R, TROEBERG L. Targeting Cartilage Degradation in Osteoarthritis. Pharmaceuticals (Basel). 2021;14(2):126.
[14] CHEN A, CHEN Y, RONG X, et al. The application of exosomes in the early diagnosis and treatment of osteoarthritis. Front Pharmacol. 2023; 14:1154135.
[15] 王刘欣,王贺林,张勉.外泌体在骨关节炎发病及治疗中的作用和机制研究[J].中国现代医学杂志,2022,32(18):63-69.
[16] QIAN Y, CHU G, ZHANG L, et al. M2 macrophage-derived exosomal miR-26b-5p regulates macrophage polarization and chondrocyte hypertrophy by targeting TLR3 and COL10A1 to alleviate osteoarthritis. J Nanobiotechnology. 2024;22(1):72.
[17] XU J, ZHOU K, GU H, et al. Exosome miR-4738-3p-mediated regulation of COL1A2 through the NF-κB and inflammation signaling pathway alleviates osteoarthritis low-grade inflammation symptoms. Biomol Biomed. 2024;24(3):520-536.
[18] QIU M, XIE Y, TAN G, et al. Synovial mesenchymal stem cell-derived exosomal miR-485-3p relieves cartilage damage in osteoarthritis by targeting the NRP1-mediated PI3K/Akt pathway: Exosomal miR-485-3p relieves cartilage damage. Heliyon. 2024;10(2):e24042.
[19] LIU Y, LU T, LIU Z, et al. Six macrophage-associated genes in synovium constitute a novel diagnostic signature for osteoarthritis. Front Immunol. 2022;13:936606.
[20] 袁长深,廖书宁,李哲,等.N6-甲基腺苷相关调节因子与骨关节炎:生物信息学和实验验证分析[J].中国组织工程研究,2024,28(11): 1724-1729.
[21] LUO D, GAO X, ZHU X, et al. Biomarker screening using integrated bioinformatics for the development of “normal-impaired glucose intolerance-type 2 diabetes mellitus”. Sci Rep. 2024;14(1):4558.
[22] WU X, BIAN B, LIN Z, et al. Identification of exosomal mRNA, lncRNA and circRNA signatures in an osteoarthritis synovial fluid-exosomal study. Exp Cell Res. 2022;410(1):112881.
[23] CHEN P, RUAN A, ZHOU J, et al. Identification and analysis of key microRNAs derived from osteoarthritis synovial fluid exosomes. Chin Med J (Engl). 2023;136(2):245-247.
[24] YI C, ZANG N, GAO L, et al. THY1 is a prognostic-related biomarker via mediating immune infiltration in lung squamous cell carcinoma (LUSC). Aging (Albany NY). 2024;16(11):9498-9517.
[25] PAINE A, WOELLER CF, ZHANG H, et al. Thy1 is a positive regulator of osteoblast differentiation and modulates bone homeostasis in obese mice. FASEB J. 2018;32(6):3174-3183.
[26] GUO HL, CHEN G, SONG ZL, et al. COL6A3 promotes cellular malignancy of osteosarcoma by activating the PI3K/AKT pathway. Rev Assoc Med Bras (1992). 2020;66(6):740-745.
[27] WANG K, PENG X, ZHANG R, et al. COL6A3 enhances the osteogenic differentiation potential of BMSCs by promoting mitophagy in the osteoporotic microenvironment. Mol Biol Rep. 2024;51(1):206.
[28] WINSLOW S, SCHOLZ A, RAPPL P, et al. Macrophages attenuate the transcription of CYP1A1 in breast tumor cells and enhance their proliferation. PLoS One. 2019;14(1):e0209694.
[29] WANG X, WU H, ZHANG Q, et al. NFKB2 inhibits NRG1 transcription to affect nucleus pulposus cell degeneration and inflammation in intervertebral disc degeneration. Mech Ageing Dev. 2021;197:111511.
[30] WEI Y, LUO L, GUI T, et al. Targeting cartilage EGFR pathway for osteoarthritis treatment. Sci Transl Med. 2021;13(576):eabb3946.
[31] LI K, ZHANG Y, ZHANG Y, et al. Tyrosine kinase Fyn promotes osteoarthritis by activating the β-catenin pathway. Ann Rheum Dis. 2018;77(6):935-943.
[32] FELSON DT, MISRA D, LAVALLEY M, et al. MOST Study Investigators. Essential Fatty Acids and Osteoarthritis. Arthritis Care Res (Hoboken). 2024;76(6):796-801.
[33] LIU J, JIA S, YANG Y, et al. Exercise induced meteorin-like protects chondrocytes against inflammation and pyroptosis in osteoarthritis by inhibiting PI3K/Akt/NF-κB and NLRP3/caspase-1/GSDMD signaling. Biomed Pharmacother. 2023;158:114118.
[34] BAN Y, WANG Y, QIAO L, et al. Total lignans from Vitex negundo seeds attenuate osteoarthritis and their main component vitedoin A alleviates osteoclast differentiation by suppressing ERK/NFATc1 signaling. Phytother Res. 2023;37(4):1422-1434.
[35] HUANG Y, LIAO J, VLASHI R, et al. Focal adhesion kinase (FAK): its structure, characteristics, and signaling in skeletal system. Cell Signal. 2023;111:110852. |