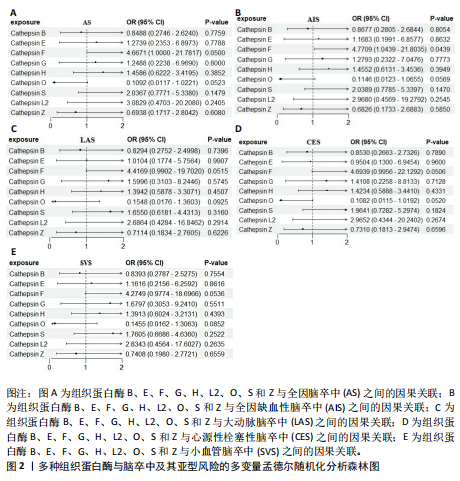
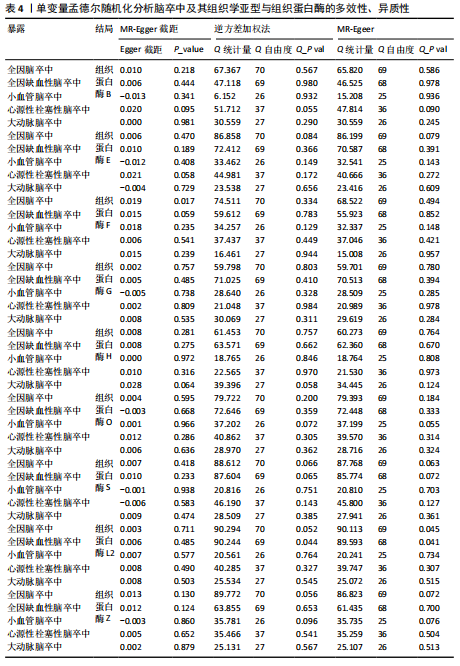
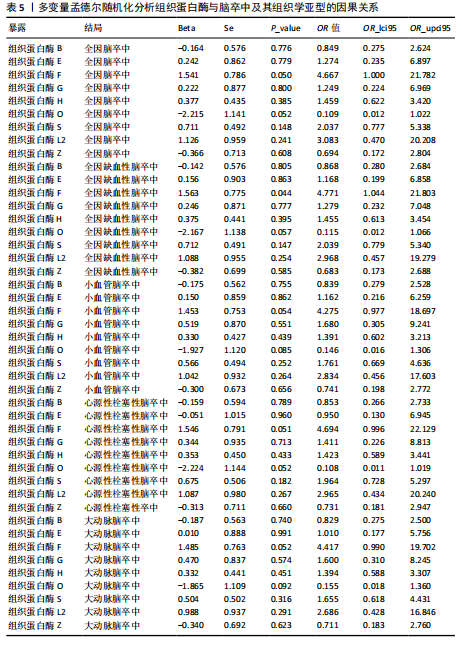
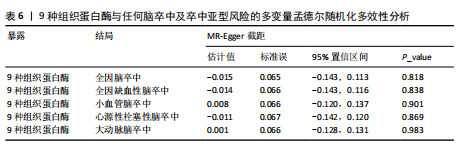
[1] GBD 2016 LIFETIME RISK OF STROKE COLLABORATORS, FEIGIN VL, NGUYEN G, et al. Global, Regional, and Country-Specific Lifetime Risks of Stroke, 1990 and 2016. N Engl J Med. 2018;379(25):2429-2437.
[2] GBD 2016 STROKE COLLABORATORS. Global, regional, and national burden of stroke, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18(5):439-458.
[3] LINDSAY MP, NORRVING B, SACCO RL, et al. World Stroke Organization (WSO): Global Stroke Fact Sheet 2019. Int J Stroke. 2019;14(8):806-817.
[4] KAHL A, BLANCO I, JACKMAN K, et al. Publisher Correction: Cerebral ischemia induces the aggregation of proteins linked to neurodegenerative diseases. Sci Rep. 2018;8(1):6802.
[5] GIFFARD RG, XU L, ZHAO H, et al. Chaperones, protein aggregation, and brain protection from hypoxic/ischemic injury. J Exp Biol. 2004;207(Pt 18):3213-3220.
[6] LIU Y, XUE X, ZHANG H, et al. Neuronal-targeted TFEB rescues dysfunction of the autophagy-lysosomal pathway and alleviates ischemic injury in permanent cerebral ischemia. Autophagy. 2019;15(3):493-509.
[7] REISER J, ADAIR B, REINHECKEL T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest. 2010;120(10):3421-3431.
[8] PATEL S, HOMAEI A, EL-SEEDI HR, et al. Cathepsins: Proteases that are vital for survival but can also be fatal. Biomed Pharmacother. 2018;105:526-532.
[9] HU K, GAIRE BP, SUBEDI L, et al. Interruption of Endolysosomal Trafficking After Focal Brain Ischemia. Front Mol Neurosci. 2021; 14:719100.
[10] YUAN D, HU K, LOKE CM, et al. Interruption of endolysosomal trafficking leads to stroke brain injury. Exp Neurol. 2021;345:113827.
[11] CARLONI S, BUONOCORE G, BALDUINI W. Protective role of autophagy in neonatal hypoxia-ischemia induced brain injury. Neurobiol Dis. 2008;32(3):329-339.
[12] WEN YD, SHENG R, ZHANG LS, et al. Neuronal injury in rat model of permanent focal cerebral ischemia is associated with activation of autophagic and lysosomal pathways. Autophagy. 2008;4(6):762-769.
[13] RAMI A, LANGHAGEN A, STEIGER S. Focal cerebral ischemia induces upregulation of Beclin 1 and autophagy-like cell death. Neurobiol Dis. 2008;29(1):132-141.
[14] ZHANG X, YAN H, YUAN Y, et al. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013;9(9):1321-1333.
[15] Nakanishi H. Microglial cathepsin B as a key driver of inflammatory brain diseases and brain aging. Neural Regen Res. 2020; 15(1):25-29.
[16] BRENNAN P, HAINAUT P, BOFFETTA P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12(4):399-408.
[17] JULIAN TH, BODDY S, ISLAM M, et al. A review of Mendelian randomization in amyotrophic lateral sclerosis. Brain. 2022;145(3):832-842.
[18] SUN BB, MARANVILLE JC, PETERS JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73-79.
[19] MALIK R, CHAUHAN G, TRAYLOR M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537.
[20] LIN Z, DENG Y, PAN W. Combining the strengths of inverse-variance weighting and Egger regression in Mendelian randomization using a mixture of regressions model. PLoS Genet. 2021; 17(11):e1009922.
[21] BOWDEN J, DAVEY SMITH G, HAYCOCK PC, et al. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304-314.
[22] BOWDEN J, DAVEY SMITH G, BURGESS S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525.
[23] COHEN JF, CHALUMEAU M, COHEN R, et al. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J Clin Epidemiol. 2015;68(3):299-306.
[24] Gala H, Tomlinson I. The use of Mendelian randomisation to identify causal cancer risk factors: promise and limitations. J Pathol. 2020;250(5):541-554.
[25] HILKENS NA, CASOLLA B, LEUNG TW, et al. Stroke. Lancet. 2024;403(10446):2820-2836.
[26] EKKER MS, VERHOEVEN JI, VAARTJES I, et al. Stroke incidence in young adults according to age, subtype, sex, and time trends. Neurology. 2019;92(21):e2444-e2454.
[27] YIIN GS, HOWARD DP, PAUL NL, et al. Age-specific incidence, outcome, cost, and projected future burden of atrial fibrillation-related embolic vascular events: a population-based study. Circulation. 2014;130(15):1236-1244.
[28] BOGIATZI C, HACKAM DG, MCLEOD AI, et al. Secular trends in ischemic stroke subtypes and stroke risk factors. Stroke. 2014;45(11):3208-3213.
[29] FERNLUND E, GYLLENHAMMAR T, JABLONOWSKI R, et al. Serum Biomarkers of Myocardial Remodeling and Coronary Dysfunction in Early Stages of Hypertrophic Cardiomyopathy in the Young. Pediatr Cardiol. 2017;38(4):853-863.
[30] CUVELLIEZ M, VANDEWALLE V, BRUNIN M, et al. Circulating proteomic signature of early death in heart failure patients with reduced ejection fraction. Sci Rep. 2019;9(1):19202.
[31] STYPMANN J, GLÄSER K, ROTH W, et al. Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin L. Proc Natl Acad Sci U S A. 2002; 99(9):6234-6239.
[32] TANG Q, CAI J, SHEN D, et al. Lysosomal cysteine peptidase cathepsin L protects against cardiac hypertrophy through blocking AKT/GSK3beta signaling. J Mol Med (Berl). 2009;87(3):249-260.
[33] BEVAN S, TRAYLOR M, ADIB-SAMII P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43(12):3161-3167.
[34] OLSON OC, JOYCE JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15(12):712-729.
[35] LIN Z, ZHAO S, LI X, et al. Cathepsin B S-nitrosylation promotes ADAR1-mediated editing of its own mRNA transcript via an ADD1/MATR3 regulatory axis. Cell Res. 2023;33(7):546-561.
[36] LI J, CHEN Z, KIM G, et al. Cathepsin W restrains peripheral regulatory T cells for mucosal immune quiescence. Sci Adv. 2023;9(28):eadf3924.
[37] ARORA K, HERROON M, AL-AFYOUNI MH, et al. Catch and Release Photosensitizers: Combining Dual-Action Ruthenium Complexes with Protease Inactivation for Targeting Invasive Cancers. J Am Chem Soc. 2018;140(43):14367-14380.
[38] FRUGÉ AD, SMITH KS, BAIL JR, et al. Biomarkers Associated With Tumor Ki67 and Cathepsin L Gene Expression in Prostate Cancer Patients Participating in a Presurgical Weight Loss Trial. Front Oncol. 2020;10:544201.
[39] BARARIA D, HILDEBRAND JA, STOLZ S, et al.Cathepsin S Alterations Induce a Tumor-Promoting Immune Microenvironment in Follicular Lymphoma. Cell Rep. 2020; 31(5):107522.
[40] SONG L, WANG X, CHENG W, et al. Expression signature, prognosis value and immune characteristics of cathepsin F in non-small cell lung cancer identified by bioinformatics assessment. BMC Pulm Med. 2021;21(1):420.
[41] WEI S, LIU W, XU M, et al. Cathepsin F and Fibulin-1 as novel diagnostic biomarkers for brain metastasis of non-small cell lung cancer. Br J Cancer. 2022;126(12):1795-1805.
[42] SANTAMARÍA I, VELASCO G, PENDÁS AM, et al. Molecular cloning and structural and functional characterization of human cathepsin F, a new cysteine proteinase of the papain family with a long propeptide domain. J Biol Chem. 1999;274(20): 13800-13809.
[43] ZDRAVKOVA K, MIJANOVIC O, BRANKOVIC A, et al. Unveiling the Roles of Cysteine Proteinases F and W: From Structure to Pathological Implications and Therapeutic Targets. Cells. 2024;13(11):917.
[44] HSU A, PODVIN S, HOOK V. Lysosomal Cathepsin Protease Gene Expression Profiles in the Human Brain During Normal Development. J Mol Neurosci. 2018;65(4):420-431.
[45] TANG CH, LEE JW, GALVEZ MG, et al. Murine cathepsin F deficiency causes neuronal lipofuscinosis and late-onset neurological disease. Mol Cell Biol. 2006;26(6):2309-2316.
[46] DALTON JP, ROBINSON MW, BRINDLEY PJ. Cathepsin// BARRETT AJ, RAWLINGS ND, WOESSNER JF, eds. Handbook of Proteolytic Enzymes. 3rd ed. Cambridge, MA: Academic Press; 2013.
[47] FINN RD, BATEMAN A, CLEMENTS J, et al.Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue): D222-D230.
[48] DI FABIO R, MORO F, PESTILLO L, et al. Pseudo-dominant inheritance of a novel CTSF mutation associated with type B Kufs disease. Neurology. 2014;83(19):1769-1770. |