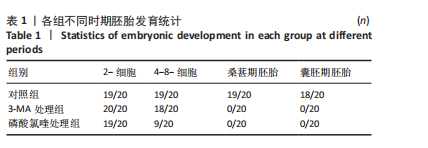
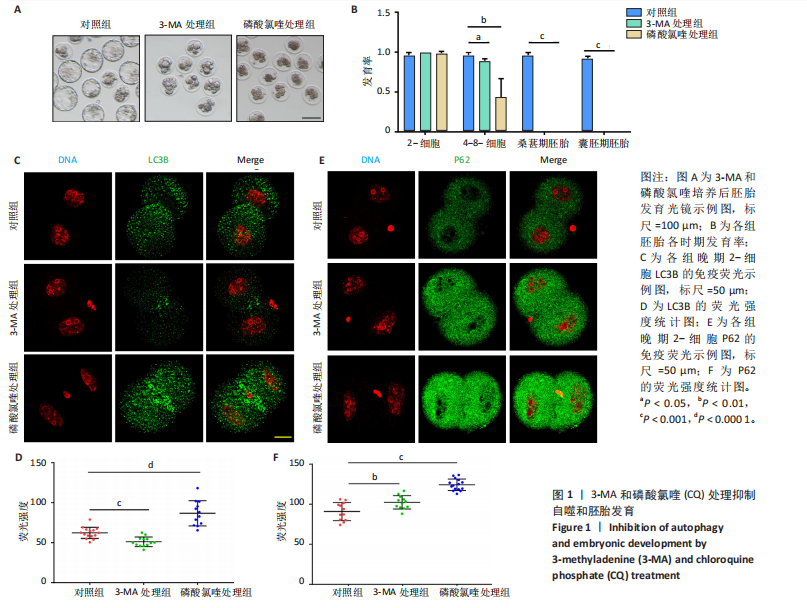
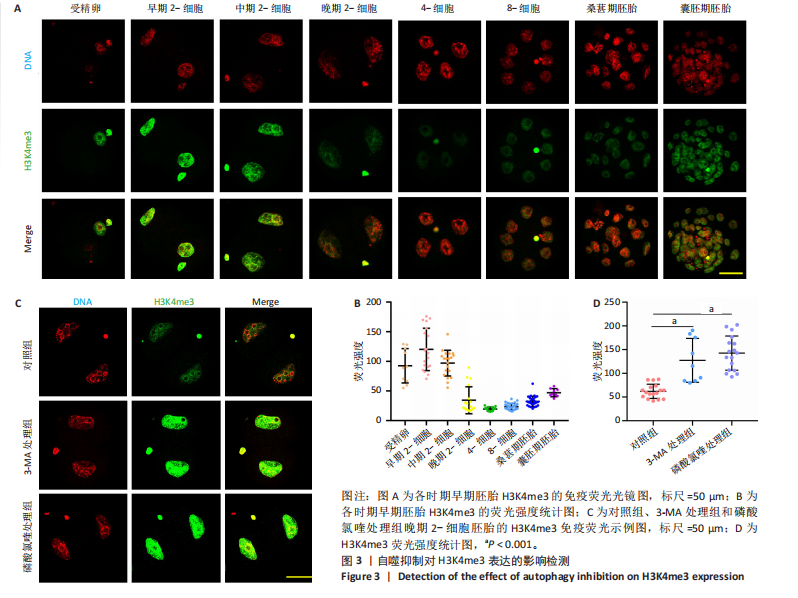
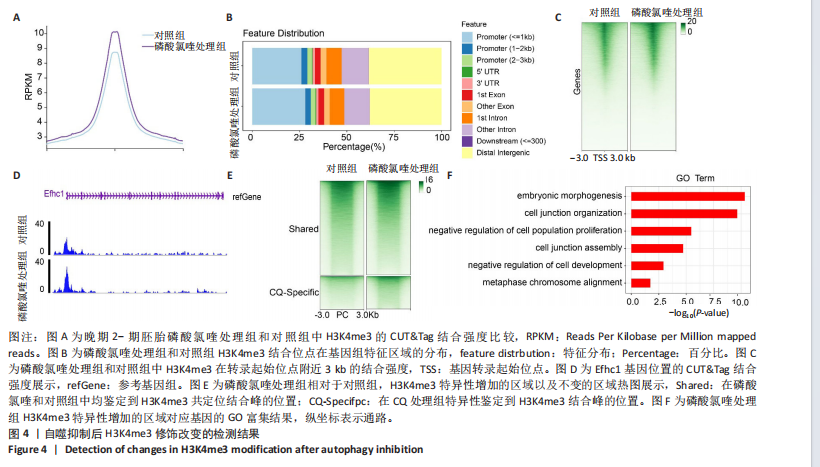
[1] FENG Y, CHEN Y, WU X, et al. Interplay of energy metabolism and autophagy. Autophagy. 2024;20(1):4-14.
[2] BOYA P, CODOGNO P, RODRIGUEZ-MUELA N. Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development. 2018;145(4):dev146506.
[3] TABIBZADEH S. Role of autophagy in aging: The good, the bad, and the ugly. Aging Cell. 2023;22(1):e13753.
[4] GÓMEZ-VIRGILIO L, SILVA-LUCERO M-DC, FLORES-MORELOS DS, et al. Autophagy: A Key Regulator of Homeostasis and Disease: An Overview of Molecular Mechanisms and Modulators. Cells. 2022;11(15):2262.
[5] ALLEN EA, BAEHRECKE EH. Autophagy in animal development. Cell Death Differ. 2020;27(3): 903-918.
[6] SONG S, GUO Q, ZHU Y, et al. Exploring the role of autophagy during early human embryonic development through single-cell transcriptome and methylome analyses. Sci China Life Sci. 2022;65(5):940-952.
[7] TSUKAMOTO S, KUMA A, MURAKAMI M, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008; 321(5885):117-120.
[8] TSUKAMOTO S, KUMA A, MIZUSHIMA N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008;4(8):1076-1078.
[9] KOMATSU M, WAGURI S, UENO T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425-434.
[10] YUE Z, JIN S, YANG C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077-15082.
[11] HONDA BM, DIXON GH, CANDIDO EP. Sites of in vivo histone methylation in developing trout testis. J Biol Chem. 1975;250(22): 8681-8685.
[12] ALBERT TK, KERL K. A histone tale that enCOMPASSes pausing: new insights into the functional repertoire of H3K4me3. Signal Transduct Target Ther. 2023;8(1):270.
[13] YU H, LESCH B J. Functional Roles of H3K4 Methylation in Transcriptional Regulation. Mol Cell Biol. 2024;44(11):505-515.
[14] BARSOUM M, STENZEL AT, BOCHYŃSKA A, et al. Loss of the Ash2l subunit of histone H3K4 methyltransferase complexes reduces chromatin accessibility at promoters. Sci Rep. 2022;12(1):21506.
[15] DAHL J A, JUNG I, AANES H, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016; 537(7621):548-552.
[16] ZHANG B, ZHENG H, HUANG B, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537(7621):553-557.
[17] CAO X, MA T, FAN R, et al. Broad H3K4me3 Domain Is Associated with Spatial Coherence during Mammalian Embryonic Development. bioRxiv [Preprint]. 2023;11:570452.
[18] ZHOU C, HALSTEAD MM, BONNET-GARNIER A, et al. Resetting H3K4me3, H3K27ac, H3K9me3 and H3K27me3 during the maternal-to-zygotic transition and blastocyst lineage specification in bovine embryos. bioRxiv. 2022;7:486777.
[19] LIU X, WANG C, LIU W, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558-562.
[20] DANG Y, LUO L, SHI Y, et al.KDM5-mediated redistribution of H3K4me3 is required for oocyte-to-embryonic transition in cattle†. Biol Reprod. 2022;106(6):1059-1071.
[21] ARTAL-MARTINEZ DE NARVAJAS A, GOMEZ TS, ZHANG JS, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33(20):3983-3993.
[22] SHIN HJ, KIM H, OH S, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534(7608):
553-557.
[23] FÜLLGRABE J, LYNCH-DAY MA, HELDRING N, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500(7463):468-471.
[24] TSUKAMOTO S, HARA T, YAMAMOTO A, et al. Fluorescence-based visualization of autophagic activity predicts mouse embryo viability. Sci Rep. 2014;4:4533.
[25] SHEN X, ZHANG N, WANG Z, et al. Induction of autophagy improves embryo viability in cloned mouse embryos. Sci Rep. 2015;5:17829.
[26] ZHANG L, ZHANG Y, HAN Z, et al. Transcriptome Analyses Reveal Effects of Vitamin C-Treated Donor Cells on Cloned Bovine Embryo Development. Int J Mol Sci. 2019;20(11):2628.
[27] XU J, SHI P, ZHAO X, et al. Cell synchronization by Rapamycin improves the developmental competence of buffalos (Bubalus bubalis) somatic cell nuclear transfer embryos. Reprod Domest Anim. 2021;56(2):313-323.
[28] DELUAO JC, WINSTANLEY Y, ROBKER RL, et al. OXIDATIVE STRESS AND REPRODUCTIVE FUNCTION: Reactive oxygen species in the mammalian pre-implantation embryo. Reproduction. 2022;164(6):F95-F108.
[29] LEITE RF, ANNES K, ISPADA J, et al. Oxidative Stress Alters the Profile of Transcription Factors Related to Early Development on In Vitro Produced Embryos. Oxid Med Cell Longev. 2017;2017:1502489.
[30] JAMIL M, DEBBARH H, ABOULMAOUAHIB S, et al. Reactive oxygen species in reproduction: harmful, essential or both?. Zygote. 2020; 28(4):255-269.
[31] KEANE JA, EALY AD. An Overview of Reactive Oxygen Species Damage Occurring during In Vitro Bovine Oocyte and Embryo Development and the Efficacy of Antioxidant Use to Limit These Adverse Effects. Animals (Basel). 2024;14(2):330.
[32] XU Q, XIE W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 2018;28(3):237-253.
[33] WILKINSON AL, ZORZAN I, RUGG-GUNN PJ. Epigenetic regulation of early human embryo development. Cell Stem Cell. 2023;30(12): 1569-1584.
[34] 张立苹, 林郑云, 华再东, 等. 组蛋白H3K4me3甲基化修饰与哺乳动物早期胚胎发育[J]. 湖北畜牧兽医,2019,40(7):16-17+19.
[35] BU G, ZHU W, LIU X, et al. Coordination of zygotic genome activation entry and exit by H3K4me3 and H3K27me3 in porcine early embryos. Genome Res. 2022;32(8):1487-1501.
[36] LI C, ZHANG Y, SHEN J, et al. Cfp1 Controls Cardiomyocyte Maturation by Modifying Histone H3K4me3 of Structural, Metabolic, and Contractile Related Genes. Adv Sci (Weinh). 2024;11(11):e2305992.
[37] SU WC, MAO XM, LI SY, et al. DPY30 Promotes Proliferation and Cell Cycle Progression of Colorectal Cancer Cells via Mediating H3K4 Trimethylation. Int J Med Sci. 2023;20(7):901-917.
|