Chinese Journal of Tissue Engineering Research ›› 2024, Vol. 28 ›› Issue (1): 93-99.doi: 10.12307/2023.768
Previous Articles Next Articles
Single-cell RNA sequencing and the pathogenesis of intervertebral disc degeneration
Cheng Haotian, Zhao Xiaofeng, Lu Xiangdong, Zhao Yibo, Fan Zhifeng, Qi Detai, Wang Xiaonan, Zhou Runtian, Jin Xinjie, Zhao Bin
- Second Clinical Medical College of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China
-
Received:2022-10-13Accepted:2022-12-24Online:2024-01-08Published:2023-06-28 -
Contact:Zhao Bin, MD, Chief physician, Second Clinical Medical College of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China -
About author:Cheng Haotian, Master candidate, Second Clinical Medical College of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China -
Supported by:Shanxi Provincial Special Project for the Transformation and Guidance of Scientific and Technological Achievements, No. 201904D121008 (to ZB)
CLC Number:
Cite this article
Cheng Haotian, Zhao Xiaofeng, Lu Xiangdong, Zhao Yibo, Fan Zhifeng, Qi Detai, Wang Xiaonan, Zhou Runtian, Jin Xinjie, Zhao Bin. Single-cell RNA sequencing and the pathogenesis of intervertebral disc degeneration[J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 93-99.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
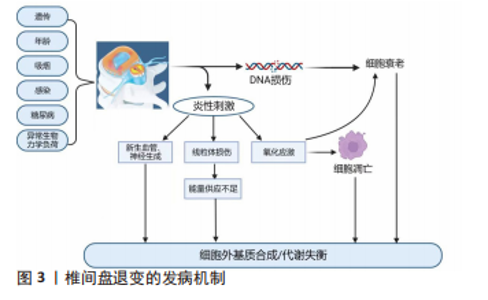
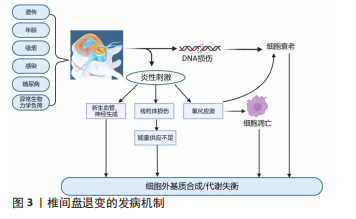
2.1 椎间盘退变概述 2.1.1 正常椎间盘结构 椎间盘是连接邻近椎体的纤维软骨组织,它通过脊柱传递由体质量和肌肉活动引起的轴向负荷,并在一定程度上维持脊柱的屈曲、伸展、侧弯和旋转等功能[12]。椎间盘由中央的髓核组织、外层的纤维环及上下软骨终板三部分组成,健康成人椎间盘中不含神经和血管。髓核组织是最重要的功能单位,在维持椎间隙高度和缓冲脊柱轴向负荷中起到关键作用[13]。髓核组织由髓核细胞和细胞外基质组成,髓核细胞负责维持细胞稳态[14],可以合成并分泌蛋白聚糖、Ⅱ型胶原等细胞外基质成分,其中Aggrecan是髓核细胞分泌最丰富的蛋白聚糖,可与透明质酸结合、聚集形成多聚体,富含带有负电荷的硫酸化基团侧链,通过调节渗透压来维持水合作用,保持髓核组织的亲水性,对脊柱承受的轴向负荷起缓冲作用。纤维环组织是由15-25个同心层组成的层状结构,可以划分为外层纤维环和内层纤维环,每层包含平行排列的Ⅰ型胶原纤维和Ⅱ型胶原纤维[15],此外弹性蛋白纤维在胶原纤维之间创建了一个跨层桥接网络,具有强大的弹性和可膨胀性,在脊柱负荷期间抵抗椎间盘的横向扩张并维持完整的结构[16]。软骨终板位于椎体和椎间盘之间,约为0.6 mm厚,结构类似于透明关节软骨,能够通过毛细血管的弥散功能将营养物质运输至椎间盘内,维持了椎间盘内的正常代谢[17]。 2.1.2 椎间盘退变发病机制 椎间盘退变是由椎间盘生理结构变化引起的,随着年龄的增长持续进展。据报道,椎间盘退变的病因可能与遗传易感性、糖尿病、年龄、吸烟、感染、异常的生物力学负荷相关[18],在影像学上表现为T2加权MRI上髓核组织的含水量减少,由高亮信号影转变为低信号黑影。根据影像学表现的不同,先后有学者依据髓核组织的信号强度、髓核组织与纤维环组织的界限及椎间隙高度分别提出了Pfirrmann分级和改良的Pfirrmann分级[19-20],用来指示椎间盘退变的发展与变化。 目前多数学者认为,椎间盘退变发生的病理生理学机制可能与多种致病因素导致的细胞外基质合成/分解代谢失衡有关,见图3,过度的机械负荷使DNA损伤,促进细胞衰老,导致转化生长因子β、胰岛素样生长因子等合成代谢因子的分泌减少,抑制了细胞外基质的合成[21],髓核细胞、纤维环细胞及免疫细胞(如巨噬细胞、T细胞、嗜中性粒细胞等)产生炎症因子,如肿瘤坏死因子α、白细胞介素1α/β、白细胞介素6、白细胞介素17、白细胞介素8、白细胞介素2、白细胞介素4、白细胞介素10、干扰素γ和各种趋化因子[22],这些炎症因子的激活诱导线粒体损伤,导致椎间盘组织能量供应不足[23],同时促进细胞外基质分解酶的合成,如基质金属蛋白酶和聚蛋白多糖酶,促进了细胞外基质的降解,引起细胞外基质代谢的不平衡[24]。此外,过度的机械负荷和炎性刺激还会诱导氧化应激,产生大量活性氧,进一步加速细胞衰老并诱导细胞凋亡[25-26]。因椎间盘组织存在于无血管、高渗、缺氧的微环境中,产生的细胞废物无法得到彻底处理,这些刺激的积累使椎间盘微环境稳态进一步失调,还会损伤组织的自我更新能力,加速椎间盘退变的进展,形成恶性循环[27]。当椎间盘组织的结构发生严重改变时,椎间隙高度大量丢失,椎间盘失去其生物力学功能,纤维环组织开始出现裂隙,大量炎症因子聚集,促进脑源性神经营养因子、神经生长因子及血管内皮生长因子生成[28],诱导椎间盘组织中新生血管形成和神经生长,这些炎性刺激的增加与神经血管生长引起的痛觉过敏导致了椎间盘源性疼痛。当纤维环组织的完整性被破坏,在长期椎间盘退变的条件下会引起椎间盘突出症、椎管狭窄症,从而出现神经压迫的症状如疼痛、麻木、无力等。"
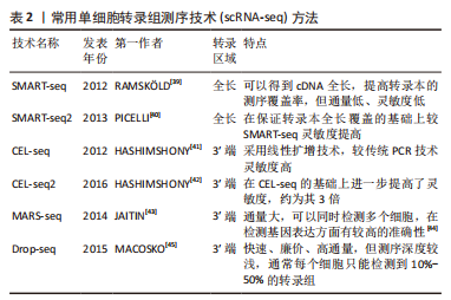
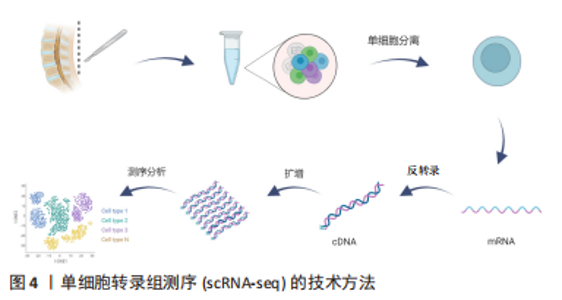
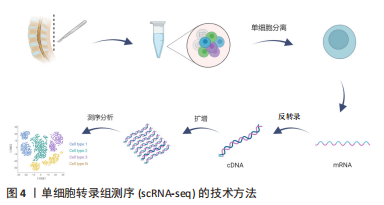
2.2.2 mRNA反转录及cDNA扩增 根据细胞类型的不同,单个细胞的RNA含量仅为1-50 pg[37],但高通量测序所需要的DNA样品量为200 ng以上,无法满足测序的要求,因此要将mRNA反转录为cDNA后进行扩增。在cDNA 扩增过程中,为避免不等比例扩增造成的测序结果偏倚,特异性分子标签(unique molecular identifiers,UMI)被当前大部分scRNA-seq技术所应用,每个UMI分别代表一个单独的转录本,使每个细胞和基因的转录本数量被量化,以校正偏倚,提高定量准确性[38]。目前常用的单细胞转录组扩增方法有模板转换法扩增技术(SMART-seq/SMART-seq2)、体外线性扩增技术(CEL-seq/CEL-seq2)、大规模平行扩增技术(MARS-seq)、微流体技术(Drop-seq),见表2。"
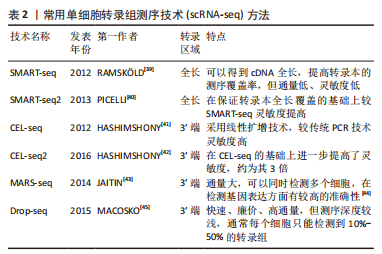
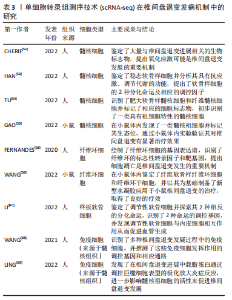
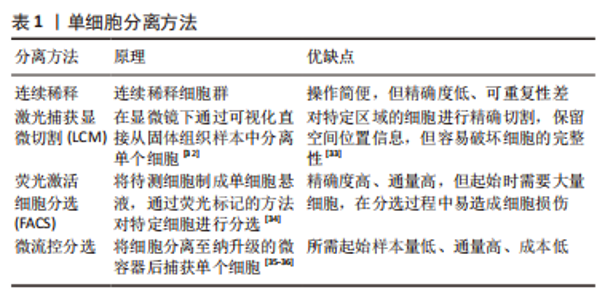
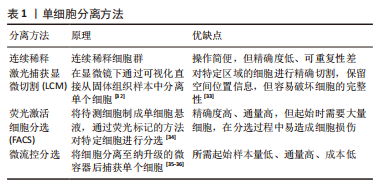
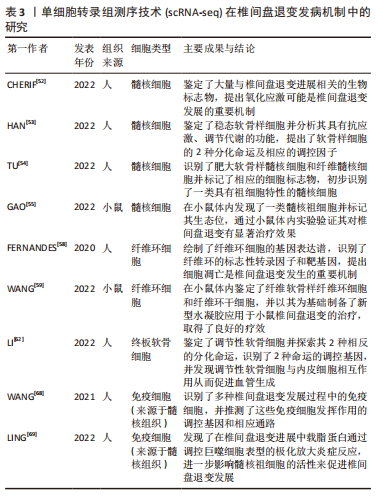
(1) SMART-seq:SMART-seq是一个具有里程碑意义的重要技术,由RAMSK?LD等[39]学者在2012年提出,在cDNA的3′末端添加几个超过模板的C碱基,再进行模板转换和PCR扩增得到cDNA全长。SMART-seq技术的优势是可以提高转录本的测序覆盖率,但具有通量低、成本高、耗时长等缺点,且灵敏度不高,在实际中应用较少。SMART-seq2是在SMART-seq的基础上优化而来,在灵敏度和准确度上有所提高,降低了扩增偏倚[40]。 (2) CEL-seq:HASHIMSHONY等[41]通过反转录带有poly-A尾巴的mRNA片段,利用体外转录(IVT)对mRNA进行线性扩增,获得足够的cDNA构建文库。这解决了以往mRNA提取量不足的问题,相比于传统的PCR扩增技术也具有更高的灵敏度。随后该研究者对CEL-seq进行改进研发出CEL-seq2,它通过使用3′端标记法显著提高了灵敏度[42],大约是CEL-seq的3倍。 (3) MARS-seq:MARS-seq是一种自动化的scRNA-seq方法,由JAITIN等[43]学者提出,通过对来自目标样本的单个细胞进行荧光激活细胞分选,可以高效地识别不同组织和疾病的独特细胞类型。MARS-seq采用3′末端计数mRNA测序方法,可生成部分cDNA转录本(非全长),在初始反转录步骤中,cDNA在体外转录(IVT)汇聚和扩增之前使用UMI标记、定量单个细胞内单个基因的表达水平,从而减少扩增步骤带来的操作偏倚,实现了cDNA扩增的多重检测,既简化了过程,又显著提高了样品通量,对复杂组织中的高效采样具有显著优势[44]。 (4) Drop-seq:MACOSKO等[45]学者开发了以微流控技术为基础的Drop-seq,他们利用微流控装置将带有UMI的oligo-dT引物和细胞一起装入微小的液滴,在各单管中平行进行反转录、PCR扩增,建立了快速、廉价、高通量的单细胞转录组测序方法,据报道其成本是荧光激活细胞分选的十分之一。但Drop-seq技术只能对3′端转录的mRNA进行测序,可能会有少量信息丢失。 2.2.3 高通量测序及平台 高通量测序技术可以对数百万个DNA同时进行测序[46],主要有全长测序与基于标签测序2种方法。目前最常用的单细胞测序平台主要是10X Genomics公司的Chromium系统和BD公司的Rhapsody系统,此外还有ICELL8 单细胞分析平台、Illumina? Bio-Rad?和Fluidigm C1单细胞全自动制备系统[47]。10xGenomics Chromium 单细胞平台的测序原理与Drop-seq技术相似,基于微流控液滴捕获细胞,能高效地进行单细胞标记、测序和分析,在短时间内每个样本可以测得几万个细胞的转录组信息,在降低成本的同时显著提高了单细胞转录组测序分析的效率。 2.2.4 数据处理及分析 scRNA-seq数据处理及分析流程包括数据预处理(质量控制、归一化、数据标准化等)、细胞和基因水平的后续分析[48]。完成高通量测序后会产生大量基础数据,在scRNA-seq数据分析前必须严格过滤数据并进行质量控制,在预处理中把特异分子标签序列、连接序列、接头序列去除,并通过移除低质量碱基和被污染的DNA等来控制数据质量,提高了数据分析的精度及效率。数据标准化是预处理的关键步骤,直接关系到下游分析结果的可靠性,目前常用的方法是CPM、Scran、Seurat和Scater等[49],通过这些统计学方法消除各单细胞文库之间的差异,降低因测序深度、表达模式不同而导致的偏倚,提高数据的可信度[50]。细胞和基因水平的后续分析是利用生物信息学手段将所得到的数据进行过滤和可视化处理,基于过滤后获得高质量的细胞基因表达谱,对细胞进行差异基因表达的鉴定、降维分析、聚类分析、细胞轨迹推测、寻找marker基因、功能富集分析等。 2.3 scRNA-seq在椎间盘退变发病机制中的研究 2.3.1 髓核及髓核细胞亚群 髓核是位于椎间盘中心的凝胶状组织,因其细胞外基质中含有大量的蛋白聚糖而具有高水平的水合作用,为椎间盘提供了独特的生物力学特性。髓核细胞是球形的软骨样细胞,分布密度约为4×106/cm2,目前被广泛认为是调控合成代谢和分解代谢并维持细胞外基质稳态的主要细胞群[51],有学者认为,髓核细胞表型和功能的变化、髓核细胞的衰老及凋亡在诱导髓核组织失水和促进椎间盘退变进展方面起着重要作用。CHERIF等[52]学者对同一个体的非退变椎间盘和退变椎间盘进行了scRNA-seq分析,基于差异基因表达的聚类分析描绘了14个不同的细胞亚群,鉴定了大量与椎间盘退变相关的生物标志物,与非退变椎间盘相比,退变椎间盘中与氧化应激反应相关的线粒体基因(MT-CYB、MT-ND2)的表达量明显下降,提示氧化应激可能是椎间盘退变发展的重要机制,此外,该团队还提出了SPTSSB、S100A1、MGP和DCN等基因通过抑制细胞外基质分解酶的活性在炎症反应、离子转运和预防组织损伤方面发挥重要作用,这些不同细胞类型及细胞亚群之间的特异性生物标志物和相互作用为椎间盘退变的治疗提出了具有潜在治疗靶点的新研究路径。 HAN等[53]通过scRNA-seq技术对正常髓核组织和退变髓核组织在基因组和转录组水平上进行分析,鉴定了稳态软骨样细胞(homeostatic chondrocytes,HomCs)并识别其上游调控因子FOS和JUN,HomCs是早期椎间盘退变细胞,功能富集于调节翻译过程及蛋白质/RNA代谢,面对拓扑不正确的蛋白质、温度异常、转化生长因子β等细胞因子的刺激,HomCs具有显著的抗应激、维持细胞稳态的功能,能够有效地延缓椎间盘退变的进展。剖析细化细胞分化轨迹,研究发现了髓核组织中软骨样细胞在椎间盘退变进展中分化的2种截然相反的命运,命运2(F2-C)中的软骨样细胞过表达多种炎性趋化因子如CXCL2和CXCL8,上调了COL1A1、COL1A2、COL3A1和MMP13多种与细胞外基质分解相关的基因,这可能与干扰素信号通路的激活、IFIT1和IFIT2等基因的上调相关,通过发挥抗增殖和促凋亡作用来促进椎间盘退变进展,并提出干扰素诱导基因的上游调控成分JAK1/STAT1可能成为下一步从分子机制上治疗椎间盘退变的新靶点。 为了深入探究髓核细胞不同亚群与椎间盘退变发生发展的相关性,TU等[54]通过对8例椎间盘退变患者髓核组织的39 372个细胞进行了无偏转录的scRNA-seq分析,根据不同功能鉴定了髓核细胞的6个亚群,发现肥大软骨样髓核细胞(hypertrophy chondrocyte-like nucleus pulposus cells,HT-CLNPs)几乎存在于椎间盘退变全程,在其亚群HT-CLNPs-Ⅱ中发现一种独特的细胞标志物CHRDL2,可以通过竞争性抑制骨形态发生蛋白来抑制软骨细胞的发育,而其亚群HT-CLNPs-Ⅰ中功能高度富集于抑制细胞凋亡、调节昼夜节律等通路,可以促进细胞外基质的合成,在椎间盘退变的进展过程中发挥潜在的保护作用,为椎间盘退变的治疗提供新的方向。纤维髓核细胞主要存在于Ⅳ级和Ⅴ级退变髓核组织中,是晚期椎间盘退变细胞,其中纤维化相关基因CRTAC1、COL3A1和MMP2表达水平显著上调,可能通过促进细胞外基质分解、加速细胞衰老及凋亡发挥功能,纤维髓核细胞还可以在白细胞介素1β介导下上调血管内皮生长因子的表达,从而促进椎间盘中的新生血管形成,这些新生血管的形成可以带来细胞因子、免疫细胞和神经生长因子,在加速椎间盘退变进展的同时引起慢性腰背痛。为探究髓核组织是否存在祖细胞(progenitor nucleus pulposus cells,ProNPCs),该学者还进行了细胞分化轨迹分析,发现在髓核和纤维环交界区域内一类高表达CD90的髓核细胞,称之为CD90+ NPCs,并通过实验验证CD90+ NPCs可以分化为成骨细胞、脂肪细胞和其他髓核细胞亚型,在轻度退变个体中,CD90+ NPCs具有较强的成软骨能力,因此可以认为CD90+ NPCs具有祖细胞特性,但对于其起源、分化及生态位还需进一步研究。 GAO等[55]通过scRNA-seq技术发现一类表达尿紧张素Ⅱ受体(urotensin Ⅱ receptor,UTS2R)的髓核细胞群对细胞迁移、生长具有调控功能,谱系示踪表明UTS2R ProNPCs以其生态位因子tenascin-C(TNC)存在于髓核组织外围,产生功能性髓核细胞,经分析参与细胞分化、发育和稳态相关的几条信号通路(如转化生长因子β、表皮细胞生长因子等),随后通过体内实验将UTS2R ProNPCs联合TNC移植到小鼠受伤的椎间盘中,结果发现治疗组可明显延缓椎间盘退变的进展,确定了具有祖细胞特性的UTS2R ProNPCs,为椎间盘退变的治疗提供新的线索和策略。这些结果为在单细胞分辨率和全转录组范围内鉴定髓核细胞亚群提供了新的见解,通过识别关键细胞亚群、祖细胞、信号通路和转录因子,模拟细胞亚群间的相互作用,深入了解细胞命运的确定,有效地为人类椎间盘退变发生发展相关的生物修复和医疗保健提供新的线索。 2.3.2 纤维环及纤维环细胞亚群 纤维环组织的承重功能由Ⅰ型胶原蛋白和细胞外基质实现[56],纤维环细胞外基质由纤维蛋白和软骨基质组成,当受到较高的机械应力时,纤维环细胞外基质的分解促进椎间盘退变发生[57],长期的生物力学变化还会导致纤维环组织的急性撕裂,进一步引起椎间盘突出症。FERNANDES等[58]以正常髓核组织与纤维环组织为样本绘制2种不同细胞群的基因表达谱,通过scRNA-seq对差异基因表达及功能富集分析,发现纤维环细胞的标志性转录因子FOXM1可以结合和激活其靶基因CENPF、CCNB1、CCNB2、KPNA2和PLK1来调节细胞周期和有丝分裂的进展,同时FOXM1也可以激活生存蛋白BIRC5,能够有效抑制细胞凋亡,在维持健康纤维环细胞数量和活力方面发挥重要功能。KDM4E可能参与髓核细胞转录组细胞外基质相关基因的调控,进一步调节细胞外基质代谢,此外正常的髓核细胞可能通过表达DKK3抑制Wnt/β-catenin信号通路来预防椎间盘退变的发生。 WANG等[59]通过scRNA-seq技术绘制了小鼠的纤维环单细胞图谱,分析了纤维环细胞的异质性,确定了共表达纤维化标志物SP7和软骨标志物Sox9的纤维软骨样纤维环细胞(fibro chondrocyte-like annulus fibrosus cells,FcAFs)以及纤维环干细胞(annulus fibrosus stem cells,AFSCs),设计了一种可以在体外诱导AFSCs产生FcAFs的纤维软骨诱导剂,根据纤维环细胞外基质的双重特征,将纤维软骨诱导剂结合到甲基化的丝素蛋白和透明质酸中制备出一种可光固化的新型复合水凝胶,将其植入到椎间盘退变小鼠模型的纤维环组织中,可以促进损伤的纤维环层状组织修复,同时还能有效地恢复软骨基质,为椎间盘退变提供了新的组织工程治疗思路。 2.3.3 终板软骨细胞 软骨终板是位于椎间盘上下方的透明软骨,是椎体和椎间盘的连接部分,其功能是通过弥散机制为椎间盘供给营养[60]。软骨终板是中央凹陷式结构,作为椎体和椎间盘之间的压力传递器,在压力传递过程中软骨终板可以分散部分压力,当过度的机械负荷使软骨终板区域受损时,相邻椎间盘的营养供应明显受限,促进了椎间盘退变的发生[61]。为探究终板软骨细胞间的异质性,LI等[62]学者通过scRNA-seq技术对人体非退变椎间盘组织中的终板软骨进行分析,鉴定了调节性软骨细胞,并通过拟时序分析研究其分化轨迹,在调节性软骨细胞的发育、分化过程中发现了2种截然不同的命运,命运Ⅰ的细胞功能富集于激活内在和外在的凋亡信号通路、蛋白质复叠、促进血管生成等功能,诱导软骨终板的退变;命运Ⅱ的细胞功能富集于促进成骨细胞分化、Ras蛋白信号转导和转录正向调节等功能,有助于软骨生长、维持稳态,而这2种命运受COL10A1、SPP1、IBSP、MT1G、MT1X、OGN等基因调控,研究学者认为维持这几种差异基因表达的平衡至关重要,一旦平衡状态被打破,可能发生不同的细胞功能和细胞命运。细胞间通讯分析还发现RegCs产生血管内皮生长因子A(VEGFA),通过旁分泌或自分泌的方式与内皮细胞中的血管内皮生长因子受体(VEGFA/KDR或VEGFA/FLT1信号通路)结合,这表明着调节性软骨细胞与内皮细胞之间的相互作用具有促血管生成的功能,进一步促进了椎间盘退变。因此,对于调节性软骨细胞更深层次的了解、靶向治疗的研究在将来可能成为减缓甚至逆转椎间盘退变的新方法。 2.3.4 免疫细胞 椎间盘已经被普遍认为是无免疫器官,有学者提出由纤维环、软骨终板组成的血液-髓核屏障将髓核组织从宿主免疫系统中分离出来,限制了免疫细胞和免疫介质进入髓核组织中引起炎症反应[63]。当血液-髓核屏障受损时,髓核组织会触发免疫反应,失去蛋白聚糖并变得纤维化[64],同时基质金属蛋白酶和炎症因子如白细胞介素1β、肿瘤坏死因子α等在椎间盘微环境中过量表达[65],据报道,这些细胞因子是由免疫细胞产生,如巨噬细胞、CD8 T细胞等,促进椎间盘组织产生炎性疼痛并加速了椎间盘退变的进展。LEE等[66]在小鼠针刺椎间盘模型的组织学分析中发现椎间盘退变组有明显的巨噬细胞浸润,这与SILVA等[67]的研究结果是一致的,即巨噬细胞可能在白细胞介素1β介导下调节蛋白聚糖和Ⅱ型胶原相关基因的表达来干扰细胞外基质合成,诱导椎间盘的退变;WANG等[68]通过scRNA-seq技术推测在椎间盘退变的病理过程中产生了一种特殊类型的免疫微环境,募集到多种类型的巨噬细胞和调节性T细胞(regulatory T cells,Tregs),这些免疫细胞通过转化生长因子β和MAPK等信号通路调节ID1、PTPRK和RAP2C基因的异常表达,从而促进椎间盘的病理改变。 LING等[69]对3例Pfirrmann Ⅱ级、Ⅲ级、Ⅳ级的人髓核组织进行scRNA-seq分析,鉴定了包括巨噬细胞、T细胞、髓系祖细胞和中性粒细胞在内的免疫细胞,功能分析显示这些免疫细胞大部分参与中性粒细胞活化、中性粒细胞脱颗粒、凋亡信号等相关通路。轨迹分析发现在椎间盘退变的进展过程中载脂蛋白诱导了M1型巨噬细胞的活化,M2型巨噬细胞的比例下降,这表明巨噬细胞M1和M2两种亚型的动态极化可能在放大炎症级联反应中发挥重要作用,通过细胞间的相互作用分析,研究者们推测核因子κB介导的巨噬细胞极化可能通过MIF信号通路诱导炎症反应来影响髓核祖细胞的活性。总体而言,这些结果表明,椎间盘退变过程中由免疫细胞介导的炎症性改变是一个复杂的过程,涉及多种细胞类型在疾病不同阶段的相互作用,了解椎间盘中的巨噬细胞积累、定位和表型转变可以提高对椎间盘退变病理生理学的了解,可能有助于确定未来的治疗靶点,见表3。"
| [1] GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1789-1858. [2] BELITSKAYA-LEVY I, CLARK JD, SHIH MC, et al. Treatment Preferences for Chronic Low Back Pain: Views of Veterans and Their Providers. J Pain Res. 2021;14:161-171. [3] SAFIRI S, KOLAHI AA, CROSS M, et al. Prevalence, Deaths, and Disability-Adjusted Life Years Due to Musculoskeletal Disorders for 195 Countries and Territories 1990-2017. Arthritis Rheumatol. 2021;73(4):702-714. [4] KRUT Z, PELLED G, GAZIT D, et al. Stem Cells and Exosomes: New Therapies for Intervertebral Disc Degeneration. Cells. 2021;10(9):2241. [5] YANG S, ZHANG F, MA J, et al. Intervertebral disc ageing and degeneration: The antiapoptotic effect of oestrogen. Ageing Res Rev. 2020;57:100978. [6] THOMPSON T, DIAS S, POULTER D, et al. Efficacy and acceptability of pharmacolo gical and non-pharmacological interventions for non-specific chronic low back pain: a protocol for a systematic review and network meta-analysis. Syst Rev. 2020;9(1):130. [7] HEINDEL P, TUCHMAN A, HSIEH PC, et al. Reoperation Rates After Single-level Lumbar Discectomy. Spine (Phila Pa 1976). 2017;42(8): E496-E501. [8] DOWDELL J, ERWIN M, CHOMA T, et al. Intervertebral Disk Degeneration and Repair. Neurosurgery. 2017;80(3S):S46-S54. [9] FILBIN MG, TIROSH I, HOVESTADT V, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360(6386):331-335. [10] SKELLY DA, SQUIERS GT, MCLELLAN MA, et al. Single-Cell Transcriptional Profiling Reveals Cellular Diversity and Intercommunication in the Mouse Heart. Cell Rep. 2018;22(3):600-610. [11] SHAH PT, STRATTON JA, STYKEL MG, et al. Single-Cell Transcriptomics and Fate Mapping of Ependymal Cells Reveals an Absence of Neural Stem Cell Function. Cell. 2018;173(4):1045-1057.e9. [12] MOLLADAVOODI S, MCMORRAN J, GREGORY D. Mechanobiology of annulus fibrosus and nucleus pulposus cells in intervertebral discs. Cell Tissue Res. 2020;379(3):429-444. [13] KHAN AN, JACOBSEN HE, KHAN J, et al. Inflammatory biomarkers of low back pain and disc degeneration: a review. Ann N Y Acad Sci. 2017;1410(1):68-84. [14] MORRIS H, GONÇALVES CF, DUDEK M, et al. Tissue physiology revolving around the clock: circadian rhythms as exemplified by the intervertebral disc. Ann Rheum Dis. 2021;80(7):828-839. [15] GHEZELBASH F, ESKANDARI AH, SHIRAZI-ADL A, et al. Modeling of human intervertebral disc annulus fibrosus with complex multi-fiber networks. Acta Biomater. 2021;123:208-221. [16] KHAN AN, JACOBSEN HE, KHAN J, et al. Inflammatory biomarkers of low back pain and disc degeneration: a review. Ann N Y Acad Sci. 2017;1410(1):68-84. [17] VADALÀ G, AMBROSIO L, RUSSO F, et al. Interaction between Mesenchymal Stem Cells and Intervertebral Disc Microenvironment: From Cell Therapy to Tissue Engineering. Stem Cells Int. 2019;2019: 2376172. [18] RISBUD MV, SHAPIRO IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10(1): 44-56. [19] PFIRRMANN CW, METZDORF A, ZANETTI M, et al. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976). 2001;26(17):1873-1878. [20] GRIFFITH JF, WANG YX, ANTONIO GE, et al. Modified Pfirrmann grading system for lumbar intervertebral disc degeneration. Spine (Phila Pa 1976). 2007;32(24):E708-E712. [21] WANG F, CAI F, SHI R, et al. Aging and age related stresses: a senescence mechanism of intervertebral disc degeneration. Osteoarthritis Cartilage. 2016;24(3):398-408. [22] HIYAMA A, SUYAMA K, SAKAI D, et al. Correlational analysis of chemokine and inflammatory cytokine expression in the intervertebral disc and blood in patients with lumbar disc disease. J Orthop Res. 2022;40(5):1213-1222. [23] SONG Y, LU S, GENG W, et al. Mitochondrial quality control in intervertebral disc degeneration. Exp Mol Med. 2021;53(7):1124-1133. [24] LI Z, CHEN X, XU D, et al. Circular RNAs in nucleus pulposus cell function and interver -tebral disc degeneration. Cell Prolif. 2019;52(6):e12704. [25] WANG B, KE W, WANG K, et al. Mechanosensitive Ion Channel Piezo1 Activated by Matrix Stiffness Regulates Oxidative Stress-Induced Senescence and Apoptosis in Human Intervertebral Disc Degeneration. Oxid Med Cell Longev. 2021;2021:8884922. [26] WANG J, XIA D, LIN Y, et al. Oxidative stress-induced circKIF18A downregulation impairs MCM7-mediated anti-senescence in intervertebral disc degeneration. Exp Mol Med. 2022;54(3): 285-297. [27] NASTO LA, ROBINSON AR, NGO K, et al. Mitochondrial-derived reactive oxygen species (ROS) play a causal role in aging-related intervertebral disc degeneration. J Orthop Res. 2013;31(7):1150-1157. [28] STEFANAKIS M, AL-ABBASI M, HARDING I, et al. Annulus fissures are mechanically and chemi cally conducive to the ingrowth of nerves and blood vessels. Spine (Phila Pa 1976). 2012;37(22):1883-1891. [29] KOLODZIEJCZYK AA, KIM JK, SVENSSON V, et al. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58(4):610-620. [30] LIU T, WU H, WU S, et al. Single-Cell Sequencing Technologies for Cardiac Stem Cell Studies. Stem Cells Dev. 2017;26(21):1540-1551. [31] MURPHY TW, ZHANG Q, NALER LB, et al. Recent advances in the use of mic roflui dic technologies for single cell analysis. Analyst. 2017; 143(1):60-80. [32] AGUILAR-BRAVO B, SANCHO-BRU P. Laser capture microdissection: techniques and applications in liver diseases. Hepatol Int. 2019;13(2): 138-147. [33] ESPINA V, WULFKUHLE JD, CALVERT VS, et al. Laser-capture microdissection. Nat Protoc. 2006;1(2):586-603. [34] CAI Y, WANG J, ZOU K. The Progresses of Spermatogonial Stem Cells Sorting Using Fluorescence-Activated Cell Sorting. Stem Cell Rev Rep. 2020;16(1):94-102. [35] ZHOU WM, YAN YY, GUO QR, et al. Microfluidics applications for high-throughput single cell sequencing. Nanobiotechnology. 2021;19(1):312. [36] ZHANG X, LI T, LIU F, et al. Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems. Mol Cell. 2019; 73(1):130-142.e5. [37] 孙睿男,王佐林.单细胞转录组测序技术进展及其在间充质干细胞研究中的应用[J].口腔颌面外科杂志,2019,29(3):174-179. [38] POTTER SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol. 2018;14(8):479-492. [39] RAMSKÖLD D, LUO S, WANG YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777-782. [40] PICELLI S, BJÖRKLUND ÅK, FARIDANI OR, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013; 10(11):1096-1098. [41] HASHIMSHONY T, WAGNER F, SHER N, et al. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2(3):666-673. [42] HASHIMSHONY T, SENDEROVICH N, AVITAL G, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. [43] JAITIN DA, KENIGSBERG E, KEREN-SHAUL H, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776-779. [44] SEE P, LUM J, CHEN J, et al. A Single-Cell Sequencing Guide for Immunologists. Front Immunol. 2018;9:2425. [45] MACOSKO EZ, BASU A, SATIJA R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202-1214. [46] HEDLUND E, DENG Q. Single-cell RNA sequencing: Technical advancements and biological applications. Mol Aspects Med. 2018;59: 36-46. [47] 周健,张子安,赵海波,等.单细胞转录组测序技术在骨关节炎中的研究进展[J].中国矫形外科杂志,2021,29(7):628-631. [48] LUECKEN MD, THEIS FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15(6):e8746. [49] STUATR T, BUTLER A, HOFFMAN P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888-1902.e21. [50] 李琳,钱四化,吕天琦,等.新一代测序技术的文库制备方法研究进展[J].应用化学,2021,38(1):11-23. [51] SILAGI ES, SHAPIRO IM, RISBUD MV. Glycosaminoglycan synthesis in the nucleus pulposus: Dysregulation and the pathogenesis of disc degeneration. Matrix Biol. 2018;71-72:368-379. [52] CHERIF H, MANNARINO M, PACIS AS, et al. Single-Cell RNA-Seq Analysis of Cells from Deg enerating and Non-Degenerating Intervertebral Discs from the Same Individual Reveals New Biomarkers for Intervertebral Disc Degeneration. Int J Mol Sci. 2022;23(7):3993. [53] HAN S, ZHANG Y, ZHANG X, et al. Single-Cell RNA Sequencing of the Nucleus Pulposus Reveals Chondrocyte Differentiation and Regulation in Intervertebral Disc Degeneration. Front Cell Dev Biol. 2022;10:824771. [54] TU J, LI W, YANG S, et al. Single-Cell Transcriptome Profiling Reveals Multicellular Ecosys tem of Nucleus Pulposus during Degeneration Progression. Adv Sci (Weinh). 2022;9(3):e2103631. [55] GAO B, JIANG B, XING W, et al. Discovery and Application of Postnatal Nucleus Pulposus Progenitors Essential for Intervertebral Disc Homeostasis and Degeneration. Adv Sci (Weinh). 2022;9(13):e2104888. [56] TAVAKOLI J, DIWAN AD, TIPPER JL. Advanced Strategies for the Regeneration of Lumbar Disc Annulus Fibrosus. Int J Mol Sci. 2020; 21(14):4889. [57] CHU G, SHI C, WANG H, et al. Strategies for Annulus Fibrosus Regeneration: From Biologic al Therapies to Tissue Engineering. Front Bioeng Biotechnol. 2018;6:90. [58] FERNANDES LM, KHAN NM, TROCHEZ CM, et al. Single-cell RNA-seq identifies unique transcr iptional landscapes of human nucleus pulposus and annulus fibrosus cells. Sci Rep. 2020;10(1):15263. [59] WANG H, WANG D, LUO B, et al. Decoding the annulus fibrosus cell atlas by scRNA-seq to develop an inducible composite hydrogel: A novel strategy for disc reconstruction. Bioact Mater. 2022;14:350-363. [60] 张艳琳,黄国付,邹璟,等.终板软骨细胞衰老在腰椎间盘退变中的研究进展[J].医学研究杂志,2022,51(7):173-176,134. [61] XIAO L, NI C, SHI J, et al. Analysis of Correlation Between Vertebral Endplate Change and Lumbar Disc Degeneration. Med Sci Monit. 2017;23:4932-4938. [62] LI W, ZHANG S, ZHAO Y, et al. Revealing the Key MSCs Niches and Pathogenic Genes in Influencing CEP Homeostasis: A Conjoint Analysis of Single -Cell and WGCNA. Front Immunol. 2022;13:933721. [63] SUN Z, LIU B, LUO ZJ. The Immune Privilege of the Intervertebral Disc: Implications for Intervertebral Disc Degeneration Treatment. Int J Med Sci. 2020;17(5):685-692. [64] BRIDGEN DT, FEARING BV, JING L, et al. Regulation of human nucleus pulposus cells by peptide -coupled substrates. Acta Biomater. 2017;55: 100-108. [65] WANG Y, CHE M, XIN J, et al. The role of IL-1β and TNF-α in intervertebral disc degenera tion. Biomed Pharmacother. 2020;131: 110660. [66] LEE S, MILLECAMPS M, FOSTER DZ, et al. Long-term histological analysis of innervation and macrophage infiltration in a mouse model of intervertebral disc injury-induced low back pain. Orthop Res. 2020; 38(6):1238-1247. [67] SILVA AJ, FERREIRA JR, CUNHA C, et al. Macrophages Down-Regulate Gene Expression of Intervertebral Disc Degenerative Markers Under a Pro-inflammatory Microenvironment. Front Immunol. 2019;10:1508. [68] WANG L, HE T, LIU J, et al. Revealing the Immune Infiltration Landscape and Identifying Diagnostic Biomarkers for Lumbar Disc Herniation. Front Immunol. 2021;12:666355. [69] LING Z, LIU Y, WANG Z, et al. Single-Cell RNA-Seq Analysis Reveals Macrophage Involved in the Progression of Human Intervertebral Disc Degeneration. Front Cell Dev Biol. 2022;9:833420. [70] DIAZ-HERNANDEZ ME, KHAN NM, TROCHEZ CM, et al. Derivation of notochordal cells from human embryonic stem cells reveals unique regulatory networks by single cell-transcriptomics. J Cell Physiol. 2020;235(6):5241-5255. |
| [1] | Long Qingxi, Zhang Pingshu, Liu Qing, Ou Ya, Zhang Lili, Yuan Xiaodong. Single-cell RNA sequencing reveals the heterogeneity of astrocytes [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 139-146. |
| [2] | Cao Sheng, Kong Lingwei, Xu Kun, Sun Zhijie. Correlation of cervical sagittal force line parameters with degenerative segment and Pfirrmann classification in patients with cervical intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(9): 1319-1324. |
| [3] | Lian Shilin, Zhang Yan, Jiang Qiang, Zhang Hanshuo, Li Tusheng, Ding Yu. Interventional effects of whole blood and platelet-rich plasma with different preparation methods on nucleus pulposus cells [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(8): 1199-1204. |
| [4] | Song Hehua, Wei Zairong. Diabetic peripheral neuropathy: research and therapy [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(8): 1278-1285. |
| [5] | Liu Hongwen, Li Jiao, Xu Wenhao, Nie Hua, Liu Shaojiang, Xu Jie, Yin Li. Differential expression profiles of microRNAs in muscle tissue of denervated skeletal muscle atrophy rats [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(5): 732-737. |
| [6] | Xiong Zhilin, Sun Hong, Liu Miao, Zhuang Yong. Roles of ferroptosis in intervertebral disc degeneration and osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(36): 5884-5890. |
| [7] | Sun Xiaoxian, Bai Xue, Liu Mengmin, Guo Yang, Lin Shun, Wu Wenxuan, Ma Yong, Liu Jintao. Establishing a rat model of intervertebral disc degeneration by castration of both upper limbs combined with intervertebral disc puncture [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(35): 5616-5621. |
| [8] | Pan Yujun, Shi Changjiang, Cao Sheng, Mu Huaizhao, Zhang Yilong, Wang Kaihua. An injectable dextran/gelatin composite hydrogel loaded with exosomes for repair of rat intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(34): 5477-5482. |
| [9] | Zhang Shijing, Huang Zhuojian, Tao Quyuan, Hou Jiangtao, Peng Bin, Luo Yongxin, Li Yanwu, Li Huibiao, Zhong Jiamin, Chen Xinlin. Molecular characteristic of immune cells in ulcerative colitis patients with spleen deficiency and dampness syndrome based on single-cell RNA sequencing [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(33): 5357-5362. |
| [10] | Wu Hao, Ye Dongping. Application of single-cell RNA sequencing in intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(33): 5370-5371. |
| [11] | Yang Chaoqiang, Zhang Hulin, Wang Xiaomin, Wang Liang, Wang Yican. Prevention and treatment of bone-related diseases by regulating macrophages with Chinese medicine [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(31): 5064-5070. |
| [12] | Xu Guangming, Liang Ziyang, Wang Hongbo, Zhang Zhen, Xiao Qinghua, Yang Jiyong, Lin Xiaosheng. Mechanical effect of oblique-pulling manipulation on triarticular complex after human lumbar intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(27): 4277-4282. |
| [13] | Chen Jiali, Gao Hang, Zhao Ziying, Wang Guangyi. Effect of traditional Chinese medicine hot compress on autophagy and expression of apoptosis related factors in the intervertebral disc of rabbits with cervical spondylosis [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(26): 4181-4186. |
| [14] | Cao Zhipeng, Shi Yu, Li Ke, Lin Wenzheng, Jiang Letao, Bu Wenzhen, Zhu Hai, Du Jianwei, Wang Huihui, Chen Hao. Carbon dot Prussian blue nanoenzyme antioxidative stress delays intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(25): 4038-4044. |
| [15] | Zhang Jian, Lin Jianping, Zhou Gang, Wang Benchao, Wu Yongchang. Etiological analysis of discoid meniscus based on whole exome sequencing [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(2): 192-199. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||