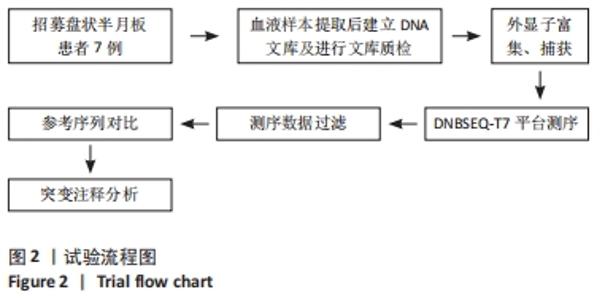
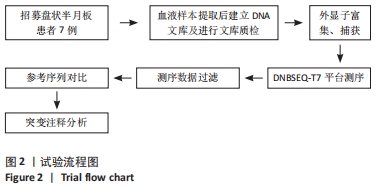
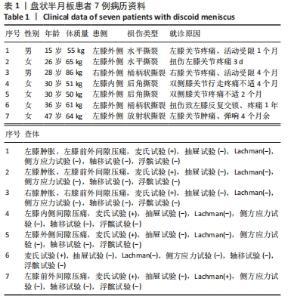
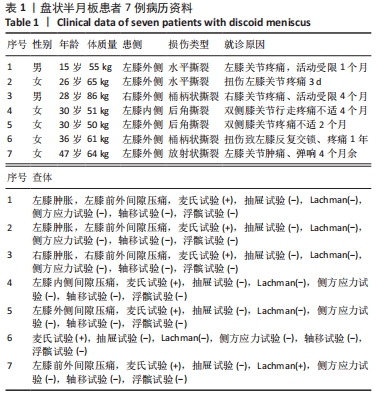
Chinese Journal of Tissue Engineering Research ›› 2023, Vol. 27 ›› Issue (2): 192-199.doi: 10.12307/2022.931
Previous Articles Next Articles
Etiological analysis of discoid meniscus based on whole exome sequencing
Zhang Jian, Lin Jianping, Zhou Gang, Wang Benchao, Wu Yongchang
- Department of Joint Surgery, Hainan Affiliated Hospital of Hainan Medical University (Hainan General Hospital), Haikou 570311, Hainan Province, China
-
Received:2021-11-18Accepted:2021-12-22Online:2023-01-18Published:2022-06-18 -
Contact:Lin Jianping, Master’s supervisor, Chief physician, Department of Joint Surgery, Hainan Affiliated Hospital of Hainan Medical University (Hainan General Hospital), Haikou 570311, Hainan Province, China -
About author:Zhang Jian, Master candidate, Physician, Department of Joint Surgery, Hainan Affiliated Hospital of Hainan Medical University (Hainan General Hospital), Haikou 570311, Hainan Province, China -
Supported by:Hainan Province Key Research and Development Task Plan, No. ZDYF2017112 (to LJP); Graduate Innovation and Entrepreneurship of Hainan Medical University, No. HYYS2020-36 (to ZJ)
CLC Number:
Cite this article
Zhang Jian, Lin Jianping, Zhou Gang, Wang Benchao, Wu Yongchang. Etiological analysis of discoid meniscus based on whole exome sequencing[J]. Chinese Journal of Tissue Engineering Research, 2023, 27(2): 192-199.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
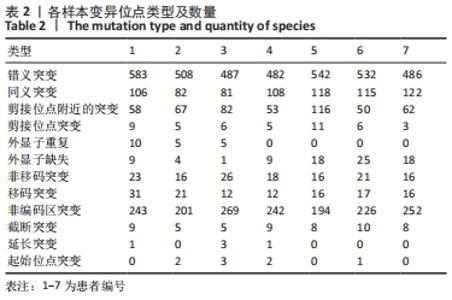
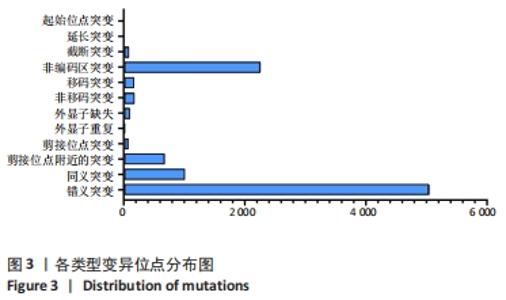
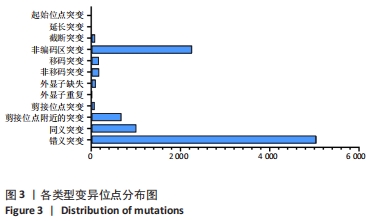
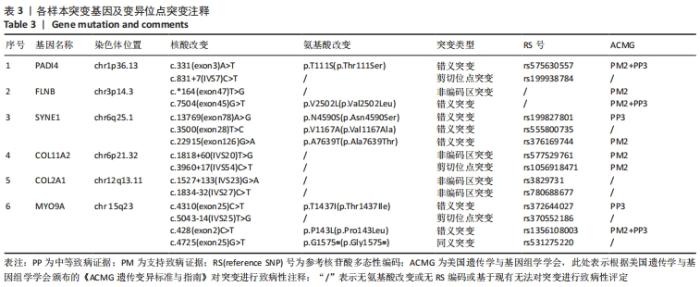
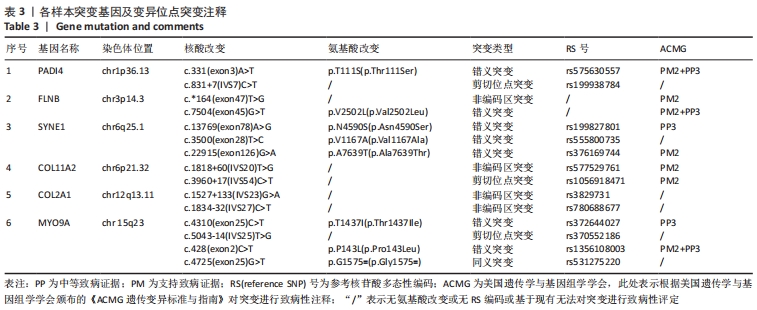
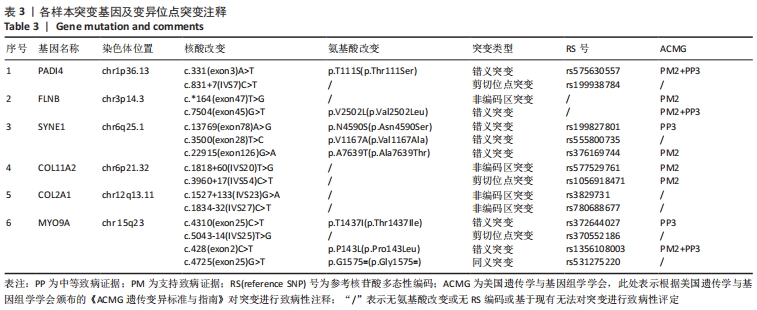
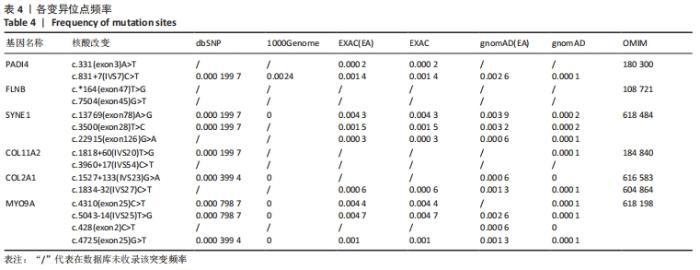
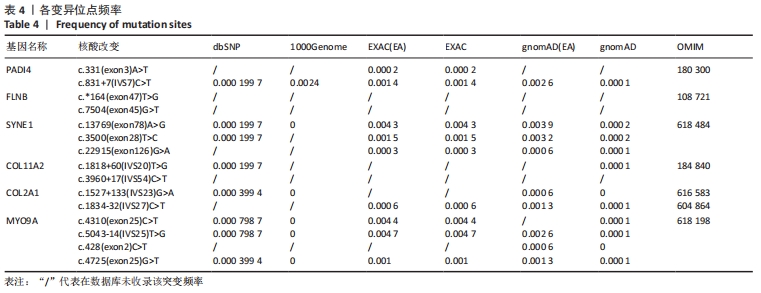
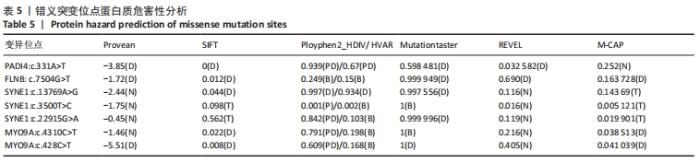
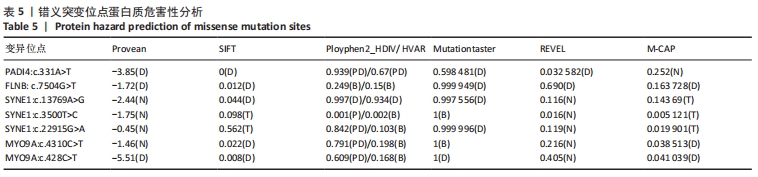
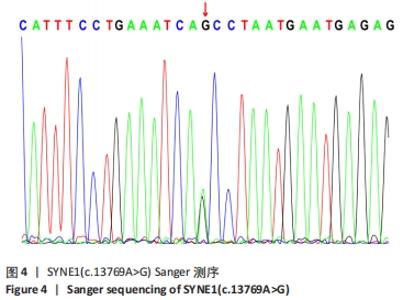
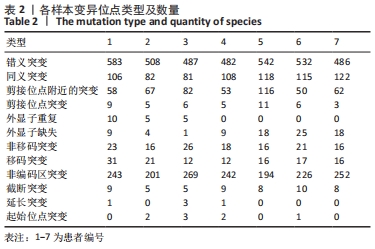
2.4 基因检测结果 通过全外显子测序检测7例盘状半月板患者外周血样本,共检测出外显子变异位点6 943个,其中错义突变3 620个,同义突变732个,剪接位点附近的突变488个,剪接位点突变45个,外显子重复20个,外显子缺失84个,非移码突变136个,截断突变54个,延长突变5个,起始位点突变8个,各样本突变情况,见表2,突变分布情况,见图3。以OMIM疾病数据库为依据,筛选骨软骨发育相关共有突变基因6个,分别为PADI4(chr1p36.13)、FLNB(chr3p14.3)、SYNE1(chr6q25.1)、COL11A2(chr6p21.32)、COL2A1(chr12q13.11)、MYO9A(chr15q23),6个共有突变基因所含变异位点共15个,分别为PADI4[c.331(exon3)A>T、c.831+7(IVS7)C>T]、FLNB[c*164(exon47)T>G、c.7504(exon45)G>T]、SYNE1[c.13769 (exon78)A>G、c.3500(exon28)T>C、c.22915(exon126)G>A]、COL11A2[c.1818+60(IVS20)T>G、c.3960+17(IVS54)C>T]、COL2A1[c.1527+133(IVS23)G>A、c.1834-32(IVS27)C>T]、MYO9A[c.4310(exon25)C>T、c.428(exon2)C>T、c.5043-14(IVS25)T>G、c.4725(exon25)G>T],其中错义突变7个,非编码区突变4个,剪切位点突变3个,同义突变1个,致病证据为PM2+PP3的变异位点为PADI4[c.331(exon3)A>T]、FLNB[c.7504(exon45)G>T]和MYO9A [c.428(exon2)C>T],致病证据为PM2的变异位点为FLNB[c.*164(exon47)T>G]、COL11A2[c.1818+60(IVS20)T>G、c.3960+17(IVS54)C>T],致病证据为PP3的变异位点为SYNE1[c.13769(exon78)A>G]和MYO9A[c.4310(exon25)C>T],各变异位点详情,见表3。根据dbSNP、千人基因组、EXAC、genomeAD数据库对所筛选出的17个变异位点的发生频率进行检索,发现上述变异位点在目前已报道的人群中携带频率均较低,在东亚人群中的发生率较高的变异位点为SYNE1[c.13769(exon78)A>G]、MYO9A[c.4310(exon25)C>T、c.5043-14(IVS25)T>G],以上3个位点在EXAC(EA)中的频率分别为0.004 3,0.004 4,0.004 7,而位于FLNB基因的频率则在各个数据库中均未报道,变异频率,见表4。针对所筛选的错义突变变异位点进行蛋白质有害性预测及有害性分析,在多个蛋白质预测分析软件中均提示有害的变异位点为PADI4:c.331(exon3)A>T、FLNB:c.7504(exon45)G>T、MYO9A:c.428(exon2)C>T,其上述位点蛋白质危害性评分提示危害性较大,见表5。结合全外显子测序和生物信息学分析结果,选取PADI4(c.331A>T),FLNB(c.7504G>T),SYNE1(c.13769A>G,c.3500T>C,c.22915G>A),MYO9A(c.4310C>T,c.428C>T)分别进行验证,其中SYNE1(c.13769A>G)验证结果见图4。"
| [1] WATANABE M, TAKEDA S, IKEUCHI H. Atlas of arthroscopy. Tokyo: Igaku-Shoin,1978:88. [2] MASQUIJO JJ, BERNOCCO F, PORTA J. Discoid meniscus in children and adolescents: Correlation between morphology and meniscal tears. Rev Esp Cir Ortop Traumatol (Engl Ed). 2019;63(1):24-28. [3] SMILLIE IS. The congenital discoid meniscus. J Bone Joint Surg Br. 1948; 30B(4):671-682. [4] KAPLAN EB. Discoid lateral meniscus of the knee joint; nature, mechanism, and operative treatment. J Bone Joint Surg Am. 1957;39-A(1):77-87. [5] 吴柏年,陈作人,陆凯,等.胎儿半月板及胫骨髁形态观察与测量[J].浙江医学,1994,16(5):13-14. [6] AHMED ALI R, MCKAY S. Familial discoid medial meniscus tear in three members of a family: a case report and review of literature. Case Rep Orthop. 2014;2014:285675. [7] 陈峰,杨波.盘状半月板病因学研究现状与进展[J].中华骨与关节外科杂志,2016,9(1):80-83. [8] 钟宏彬. BGISEQ-500、Hiseq 4000和Hiseq X Ten三个测序平台之间外显子测序数据比较分析[D].广州:华南理工大学,2018. [9] 赵福香. 膝关节盘状半月板的MRI表现以及MRI诊断盘状半月板的标准研究[J].现代医用影像学,2019,28(5):1073-1074. [10] 王淑丽,王林森,王植.膝关节盘状半月板类型及损伤的MRI分析[J].临床放射学杂志,2004,23(1):66-69. [11] SAAVEDRA M, SEPÚLVEDA M, JESÚS TUCA M, et al. Discoid meniscus: current concepts. EFORT Open Rev. 2020;5(7):371-379. [12] KOCHER MS, LOGAN CA, KRAMER DE. Discoid Lateral Meniscus in Children: Diagnosis, Management, and Outcomes. J Am Acad Orthop Surg. 2017;25(11):736-743. [13] KIM JH, AHN JH, KIM JH, et al. Discoid lateral meniscus: importance, diagnosis, and treatment. J Exp Orthop. 2020;7(1):81. [14] CHOI YH, SEO YJ, HA JM, et al. Collagenous Ultrastructure of the Discoid Meniscus: A Transmission Electron Microscopy Study. Am J Sports Med. 2017;45(3):598-603. [15] BISICCHIA S, BOTTI F, TUDISCO C. Discoid lateral meniscus in children and adolescents: a histological study. J Exp Orthop. 2018;5(1):39. [16] AYDIN KABAKÇI AD, BÜYÜKMUMCU M, AKIN D, et al. Morphological structure and variations of fetal lateral meniscus: the significance in convenient diagnosis and treatment. Knee Surg Sports Traumatol Arthrosc. 2019;27(10):3364-3373. [17] KIM JH, BIN SI, LEE BS, et al. Does discoid lateral meniscus have inborn peripheral rim instability? Comparison between intact discoid lateral meniscus and normal lateral meniscus. Arch Orthop Trauma Surg. 2018;138(12):1725-1730. [18] YANG SJ, DING ZJ, LI J, et al. Factors influencing postoperative outcomes in patients with symptomatic discoid lateral meniscus. BMC Musculoskelet Disord. 2020;21(1):551. [19] GRIMM NL, PACE JL, LEVY BJ, et al. Demographics and Epidemiology of Discoid Menisci of the Knee: Analysis of a Large Regional Insurance Database. Orthop J Sports Med. 2020;8(9):2325967120950669. [20] ATAY OA, PEKMEZCI M, DORAL MN, et al. Discoid meniscus: an ultrastructural study with transmission electron microscopy. Am J Sports Med. 2007;35(3):475-478. [21] CUI JH, MIN BH. Collagenous fibril texture of the discoid lateral meniscus. Arthroscopy. 2007;23(6):635-641. [22] Papadopoulos A, Kirkos JM, Kapetanos GA. Histomorphologic study of discoid meniscus. Arthroscopy. 2009;25(3):262-268. [23] YANG X, SHAO D. Bilateral discoid medial Meniscus: Two case reports. Medicine (Baltimore). 2019;98(15):e15182. [24] FARLETT J, WOOD JR. Ipsilateral Medial and Lateral Discoid Menisci: A Rare Combination of Infrequent Anatomic Variants. J Clin Imaging Sci. 2020;10:11. [25] 陈佳.腰椎间盘退变和先天性脊柱侧凸相关基因关联分析和致病性研究[D].北京:北京协和医学院,2018. [26] 赵春霞,崔映宇,陈义汉.核膜血影重复蛋白研究进展[J].生物化学与生物物理进展,2014,41(4):337-344. [27] GAMA MT, HOULE G, NOREAU A, et al. SYNE1 mutations cause autosomal-recessive ataxia with retained reflexes in Brazilian patients. Mov Disord. 2016;31(11):1754-1756. [28] WIETHOFF S, HERSHESON J, BETTENCOURT C, et al. Heterogeneity in clinical features and disease severity in ataxia-associated SYNE1 mutations. J Neurol. 2016;263(8):1503-1510. [29] KIM JS, KIM AR, YOUN J, et al. Identifying SYNE1 ataxia and extending the mutational spectrum in Korea. Parkinsonism Relat Disord. 2019;58: 74-78. [30] INDELICATO E, NACHBAUER W, FAUTH C, et al. SYNE1-ataxia: Novel genotypic and phenotypic findings. Parkinsonism Relat Disord. 2019;62: 210-214. [31] WANG Y, CAO R, YANG W, et al. SP1-SYNE1-AS1-miR-525-5p feedback loop regulates Ang-II-induced cardiac hypertrophy. J Cell Physiol. 2019; 234(8):14319-14329. [32] ZHAO T, MA Y, ZHANG Z, et al. Young and early-onset dilated cardiomyopathy with malignant ventricular arrhythmia and sudden cardiac death induced by the heterozygous LDB3, MYH6, and SYNE1 missense mutations. Ann Noninvasive Electrocardiol. 2021;26(4): e12840. [33] CHEN Z, REN Z, MEI W, et al. A novel SYNE1 gene mutation in a Chinese family of Emery-Dreifuss muscular dystrophy-like. BMC Med Genet. 2017;18(1):63. [34] 陈祖芝.伴有SYNE1基因新突变的Emery-Dreifuss肌营养不良症家系基因突变特点分析[D].郑州:郑州大学,2017. [35] 何梅.人核纤层蛋白病秀丽隐杆线虫模型的建立和机制研究[D].哈尔滨:东北林业大学,2018. [36] 赵丹华,赵运涛,杨旭,等. X连锁遗传的Emery-Dreifuss肌营养不良一家系的临床、病理及基因突变特点[J].中国神经免疫学和神经病学杂志,2017,24(3):197-200. [37] HARTMAN MA, FINAN D, SIVARAMAKRISHNAN S, et al. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol. 2011;27:133-155. [38] O’CONNOR E, PHAN V, CORDTS I, et al. MYO9A deficiency in motor neurons is associated with reduced neuromuscular agrin secretion. Hum Mol Genet. 2018;27(8):1434-1446. [39] O’CONNOR E, TÖPF A, MÜLLER JS, et al. Identification of mutations in the MYO9A gene in patients with congenital myasthenic syndrome. Brain. 2016;139(Pt 8):2143-2153. [40] MCMACKEN G, WHITTAKER RG, EVANGELISTA T, et al. Congenital myasthenic syndrome with episodic apnoea: clinical, neurophysiological and genetic features in the long-term follow-up of 19 patients. J Neurol. 2018;265(1):194-203. [41] YIŞ U, BECKER K, KURUL SH, et al. Genetic Landscape of Congenital Myasthenic Syndromes From Turkey: Novel Mutations and Clinical Insights. J Child Neurol. 2017;32(8):759-765. [42] TSAI CY, HSIEH SC, LIU CW, et al. The Expression of Non-Coding RNAs and Their Target Molecules in Rheumatoid Arthritis: A Molecular Basis for Rheumatoid Pathogenesis and Its Potential Clinical Applications. Int J Mol Sci. 2021;22(11):5689. [43] BADILLO-SOTO MA, RODRÍGUEZ-RODRÍGUEZ M, PÉREZ-PÉREZ ME, et al. Potential protein targets of the peptidylarginine deiminase 2 and peptidylarginine deiminase 4 enzymes in rheumatoid synovial tissue and its possible meaning. Eur J Rheumatol. 2016;3(2):44-49. [44] BAWADEKAR M, SHIM D, JOHNSON CJ, et al. Peptidylarginine deiminase 2 is required for tumor necrosis factor alpha-induced citrullination and arthritis, but not neutrophil extracellular trap formation. J Autoimmun. 2017;80:39-47. [45] 刘玮,吴歆,徐沪济.类风湿关节炎易感基因遗传背景的研究进展[J].诊断学理论与实践,2018,17(3):347-351. [46] 叶蓓,成宇,司玉莹,等.低氧环境中PADI4促进破骨细胞分化和骨吸收功能的研究[J].现代免疫学,2021,41(5):361-367. [47] LIU C, TANG W, ZHAO H, et al. The variants at FLNA and FLNB contribute to the susceptibility of hypertension and stroke with differentially expressed mRNA. Pharmacogenomics J. 2021;21(4):458-466. [48] 徐启明. FLNB基因突变在发育性骨骼畸形中的分子生物学研究[D].北京:北京协和医学院,2018. [49] 尚小可,郑君,温鹏,等.人类与实验动物半月板解剖及超微结构的比较[J].宁夏医科大学学报,2018,40(10):1231-1234. [50] 李素萍,金玉霞,杨静,等.COL2A1基因新发突变导致先天性脊柱骨骺发育不良的遗传分析及产前诊断[J].临床检验杂志,2020, 38(12): 919-921,952. [51] 张铂彦,张悦,刘贺,等.COL2A1基因突变与Ⅱ型胶原蛋白病表型关系的研究进展[J].吉林大学学报(医学版),2020,46(3):643-648. [52] TURLO AJ, MCDERMOTT BT, BARR ED, et al. Gene expression analysis of subchondral bone, cartilage, and synovium in naturally occurring equine palmar/plantar osteochondral disease. J Orthop Res. 2021. doi: 10.1002/jor.25075. Online ahead of print. [53] LIU Y, WANG L, YANG YK, et al. Prenatal diagnosis of fetal skeletal dysplasia using targeted next-generation sequencing: an analysis of 30 cases. Diagn Pathol. 2019;14(1):76. [54] XU C, LUO S, WEI L, et al. Integrated transcriptome and proteome analyses identify novel regulatory network of nucleus pulposus cells in intervertebral disc degeneration. BMC Med Genomics. 2021;14(1):40. [55] YANG X, JIA H, XING W, et al. Genetic variants in COL11A2 of lumbar disk degeneration among Chinese Han population. Mol Genet Genomic Med. 2019;7(2):e00524. |
| [1] | Li Jiajun, Xia Tian, Liu Jiamin, Chen Feng, Chen Haote, Zhuo Yinghong, Wu Weifeng. Molecular mechanism by which icariin regulates osteogenic signaling pathways in the treatment of steroid-induced avascular necrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 780-785. |
| [2] | Gao Liping, Tang Li, Wan Lu, Liu Yan, Li Xiaobing, Zhong Wenyi. Growth and development characteristics of dental and basal arch in children with skeletal class III malocclusion [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(32): 5229-5235. |
| [3] | Liang Haoran, Zhou Xin, Yang Yanfei, Niu Wenjie, Song Wenjie, Ren Zhiyuan, Wang Xueding, Liu Yang, Duan Wangping. Pathogenesis of femoral head necrosis after internal fixation of femoral neck fractures in young adults [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(3): 456-460. |
| [4] | Luo Di, Liang Xuezhen, Liu Jinbao, Li Jiacheng, Yan Bozhao, Xu Bo, Li Gang. Difference in osteogenesis- and angiogenesis-related protein expression in femoral head samples from patients with femoral head necrosis of different etiologies [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(11): 1641-1647. |
| [5] | Liu Guanjuan, Song Na, An Zheqing, Sun Jing, Liao Jian. Metagenomics and peri-implantitis [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(34): 5511-5516. |
| [6] | Wang Chuan, Peng Wuxun. Participation and regulation of long-chain non-coding RNA on pathogenesis of avascular necrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(33): 5381-5387. |
| [7] | Fan Chongshan, Sun Mingshuai, Han Wenchao. Proinflammatory factors and matrix metalloproteinases: status and roles in the pathogenesis of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(32): 5162-5170. |
| [8] | Zhou Yuanbo, Wang Jindong. Etiology and treatment of femoral trochlear dysplasia: congenital genetic determination or stress stimulation of patella [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(24): 3908-3913. |
| [9] | Xu Qi, Zhang Chao, Ha Chengzhi, Wang Dawei. Susceptibility genes related to non-traumatic femoral head necrosis: improving detection accuracy and developing new treatment strategy [J]. Chinese Journal of Tissue Engineering Research, 2020, 24(5): 747-752. |
| [10] | Wu Libing, Xu Yangyang, He Yujie, Wang Haiyan, Gao Shang, Enhejirigala, Li Xiaohe, Li Zhijun. Anatomical etiology and clinical significance of three-dimensional digital measurement of kidney stones [J]. Chinese Journal of Tissue Engineering Research, 2020, 24(29): 4662-4666. |
| [11] | Liang Chenliang, Zhao Zhenqun, Liu Wanlin. The role of OPG/RANKL/RANK signaling pathway in the pathogenesis of giant-cell tumor of bone [J]. Chinese Journal of Tissue Engineering Research, 2020, 24(23): 3723-3729. |
| [12] | Tian Lu, Liu Bin. Effects of physical activity and exercise on metabolic syndrome [J]. Chinese Journal of Tissue Engineering Research, 2020, 24(2): 296-302. |
| [13] | Wu Kai, Liao Liqing, Li Yikai . X-ray evaluation of the location and etiology of calcaneal spurs [J]. Chinese Journal of Tissue Engineering Research, 2020, 24(18): 2906-2910. |
| [14] | Meng Wei. Individualized treatment of knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2019, 23(32): 5216-5220. |
| [15] | Sun Fengyuan, Li Zongyuan, He Xi, Yang Zongde. Application and progress of finite element analysis in scoliosis biomechanical research [J]. Chinese Journal of Tissue Engineering Research, 2019, 23(32): 5221-5226. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||