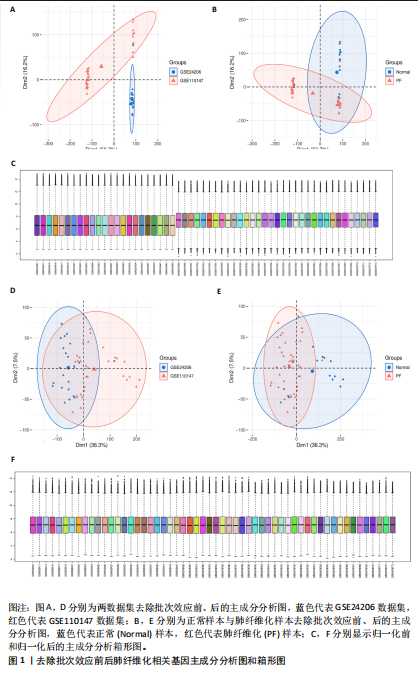
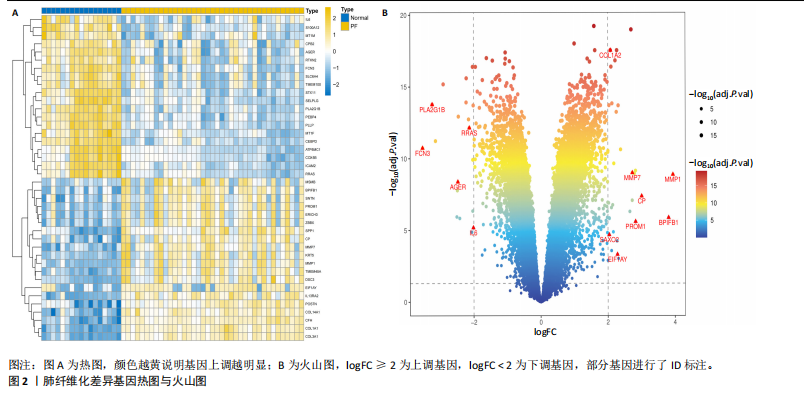
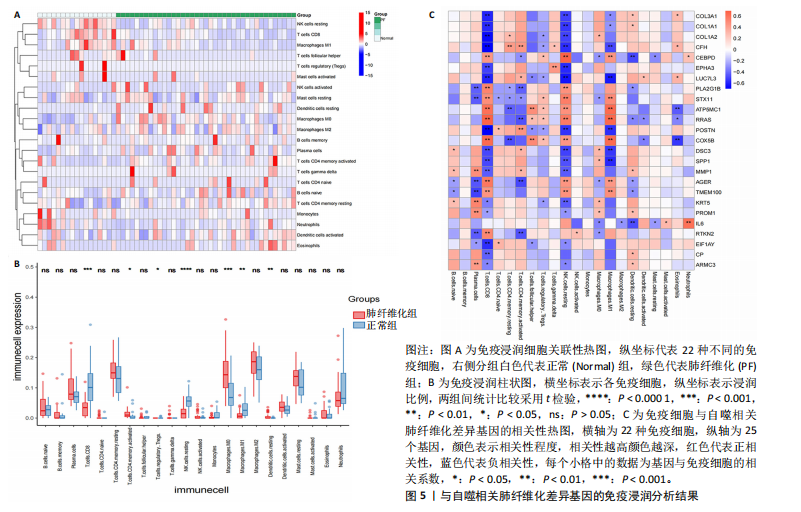
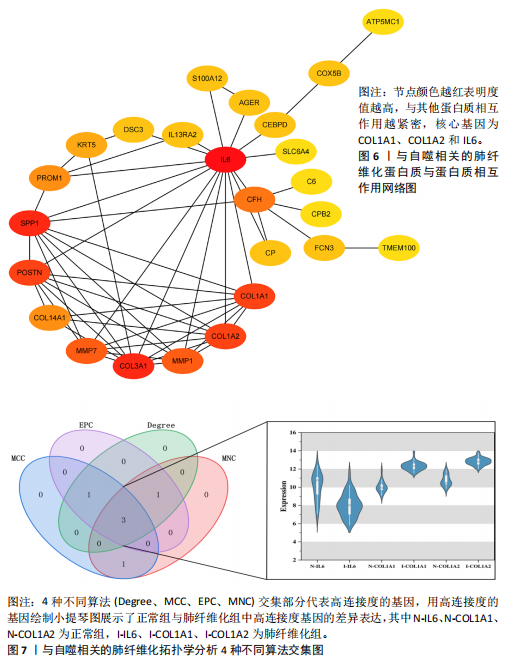
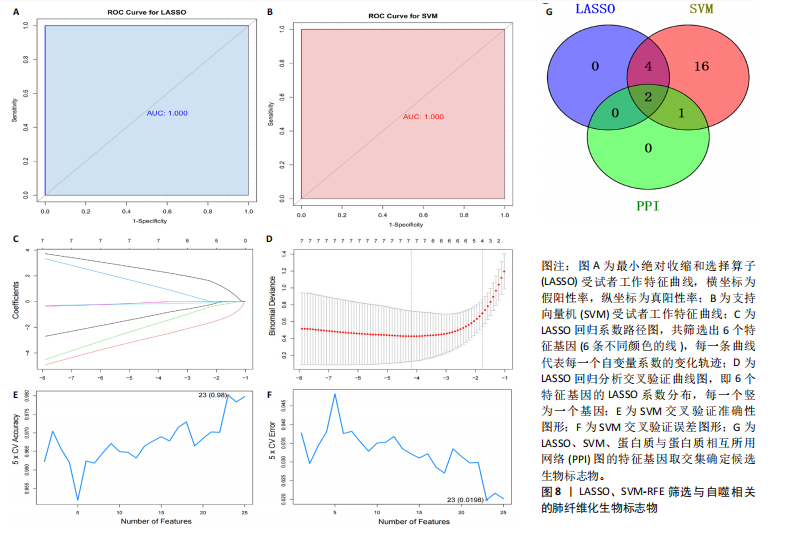
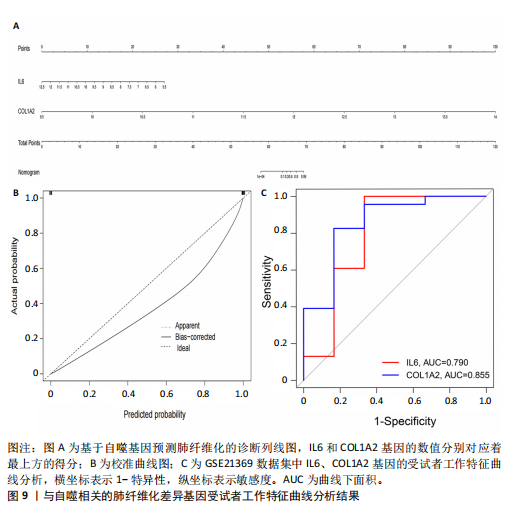
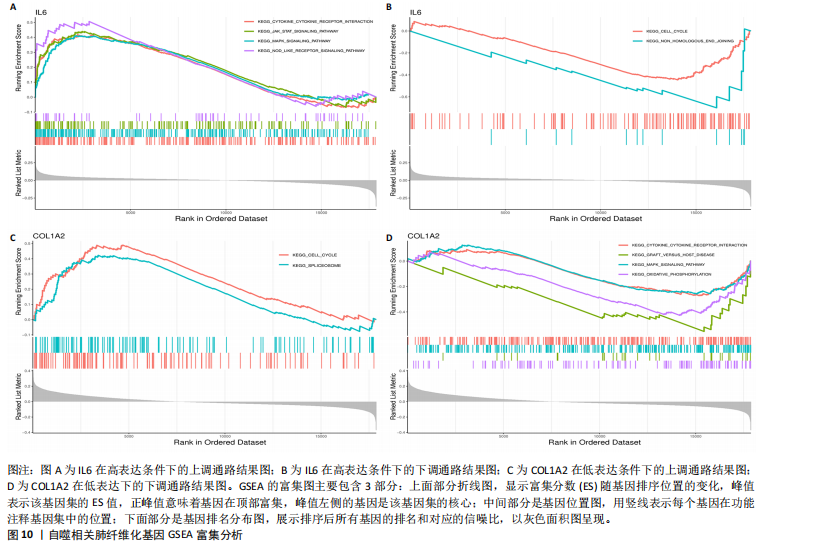
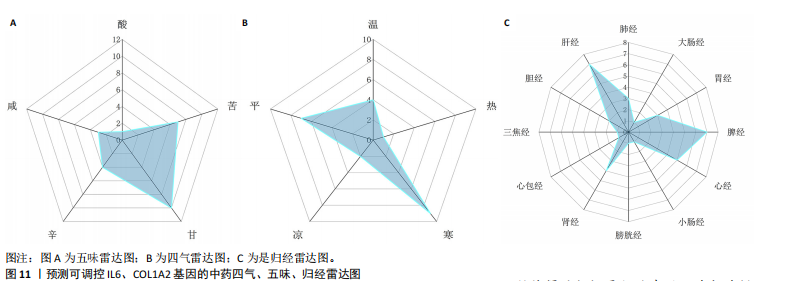
[1] ZHU JQ, TIAN YY, CHAN KL, et al. Modified Qing-Zao-Jiu-Fei decoction attenuated pulmonary fibrosis induced by bleomycin in rats via modulating Nrf2/NF-κB and MAPKs pathways. Chin Med. 2024;19(1):10.
[2] ZUO WL, ROSTAMI MR, LEBLANC M, et al. Dysregulation of club cell biology in idiopathic pulmonary fibrosis. PLoS One. 2020;15(9):e0237529.
[3] DENG Z, FEAR MW, SUK CHOI Y, et al. The extracellular matrix and mechanotransduction in pulmonary fibrosis. Int J Biochem Cell Biol. 2020;126:105802.
[4] RAGHU G, REMY-JARDIN M, RICHELDI L, et al. Idiopathic Pulmonary Fibrosis (an Update) and Progressive Pulmonary Fibrosis in Adults: An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am J Respir Crit Care Med. 2022;205(9):e18-e47.
[5] MAHER TM, BENDSTRUP E, DRON L, et al. Global incidence and prevalence of idiopathic pulmonary fibrosis. Respir Res. 2021;22(1):197.
[6] ZENG Q, JIANG D. Global trends of interstitial lung diseases from 1990 to 2019: an age-period-cohort study based on the Global Burden of Disease study 2019, and projections until 2030. Front Med (Lausanne). 2023;10:1141372.
[7] SPAGNOLO P, BALESTRO E, ALIBERTI S, et al. Pulmonary fibrosis secondary to COVID-19: a call to arms. Lancet Respir Med. 2020;8(8):750-752.
[8] COTTIN V, SCHMIDT A, CATELLA L, et al. Burden of Idiopathic Pulmonary Fibrosis Progression: A 5-Year Longitudinal Follow-Up Study. PLoS One. 2017;12(1):e0166462.
[9] PODOLANCZUK AJ, THOMSON CC, REMY-JARDIN M, et al. Idiopathic pulmonary fibrosis: state of the art for 2023. Eur Respir J. 2023;61(4):2200957.
[10] JESSEN H, HOYER N, PRIOR TS, et al. Longitudinal serological assessment of type VI collagen turnover is related to progression in a real-world cohort of idiopathic pulmonary fibrosis. BMC Pulm Med. 2021;21(1):382.
[11] KINOSHITA T, GOTO T. Molecular Mechanisms of Pulmonary Fibrogenesis and Its Progression to Lung Cancer: A Review. Int J Mol Sci. 2019;20(6):1461.
[12] SCHÄFER SC, FUNKE-CHAMBOUR M, BEREZOWSKA S. [Idiopathic pulmonary fibrosis-epidemiology, causes, and clinical course]. Pathologe. 2020;41(1):46-51.
[13] GUENTHER A, KRAUSS E, TELLO S, et al. The European IPF registry (eurIPFreg): baseline characteristics and survival of patients with idiopathic pulmonary fibrosis. Respir Res. 2018;19(1):141.
[14] JEGAL Y, PARK JS, KIM SY, et al. Clinical Features, Diagnosis, Management, and Outcomes of Idiopathic Pulmonary Fibrosis in Korea: Analysis of the Korea IPF Cohort (KICO) Registry. Tuberc Respir Dis (Seoul). 2022;85(2):185-194.
[15] Lu JL, Yu CX, Song LJ. Programmed cell death in hepatic fibrosis: current and perspectives. Cell Death Discov. 2023;9(1):449.
[16] WEN JH, LI DY, LIANG S, et al. Macrophage autophagy in macrophage polarization, chronic inflammation and organ fibrosis. Front Immunol. 2022;13:946832.
[17] ZHAN L, LI J, WEI B. Autophagy in endometriosis: Friend or foe. Biochem Biophys Res Commun. 2018;495(1):60-63.
[18] WANG K, ZHANG T, LEI Y, et al. Identification of ANXA2 (annexin A2) as a specific bleomycin target to induce pulmonary fibrosis by impeding TFEB-mediated autophagic flux. Autophagy. 2018;14(2): 269-282.
[19] ZMIJEWSKI JW, THANNICKAL VJ. Autophagy in Idiopathic Pulmonary Fibrosis: Predisposition of the Aging Lung to Fibrosis. Am J Respir Cell Mol Biol. 2023;68(1):3-4.
[20] ARAYA J, KOJIMA J, TAKASAKA N, et al. Insufficient autophagy in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2013;304(1):L56-69.
[21] CABRERA S, MACIEL M, HERRERA I, et al. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy. 2015;11(4):670-684.
[22] SHERAFATIAN M. Tree-based machine learning algorithms identified minimal set of miRNA biomarkers for breast cancer diagnosis and molecular subtyping. Gene. 2018;677:111-118.
[23] HU S, GU S, WANG S, et al. Robust Prediction of Prognosis and Immunotherapy Response for Bladder Cancer through Machine Learning Algorithm. Genes (Basel). 2022;13(6):1073.
[24] 王远敏,罗敏,苏穆,等.山柰酚对博来霉素诱导小鼠肺纤维化、MRC-5细胞胶原蛋白生成的影响[J]. 中成药,2023, 45(10):3425-3428.
[25] HERBERTS MB, TEAGUE TT, THAO V, et al. Idiopathic pulmonary fibrosis in the United States: time to diagnosis and treatment. BMC Pulm Med. 2023;23(1):281.
[26] MIGNEAULT F, HÉBERT MJ. Autophagy, tissue repair, and fibrosis: a delicate balance. Matrix Biol. 2021;100-101:182-196.
[27] MARGARITOPOULOS GA, TSITOURA E, TZANAKIS N, et al. Self-eating: friend or foe? The emerging role of autophagy in idiopathic pulmonary fibrosis. Biomed Res Int. 2013;2013:420497.
[28] MI S, LI Z, YANG HZ, et al. Blocking IL-17A promotes the resolution of pulmonary inflammation and fibrosis via TGF-beta1-dependent and -independent mechanisms. J Immunol. 2011;187(6):3003-3014.
[29] 李鹏,牛璐,赵国静,等.基于自噬探究补肺活血胶囊对博来霉素诱发小鼠肺纤维化的抑制作用[J].中成药,2024,46(4): 1337-1342.
[30] TANAKA T, NARAZAKI M, KISHIMOTO T. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy. 2016;8:959-970.
[31] PAN X, CHEN X, REN Q, et al. Single-cell transcriptomics identifies Col1a1 and Col1a2 as hub genes in obesity-induced cardiac fibrosis.Biochem Biophys Res Commun. 2022;618:30-37.
[32] 王信光,薄存香,潘志峰.IL-6家族细胞因子在肺纤维化中的作用[J].中国工业医学杂志,2024,37(3):263-266.
[33] PAPIRIS SA, TOMOS IP, KARAKATSANI A, et al. High levels of IL-6 and IL-8 characterize early-on idiopathic pulmonary fibrosis acute exacerbations. Cytokine. 2018;102:168-172.
[34] WANG Y, SANG X, SHAO R, et al. Xuanfei Baidu Decoction protects against macrophages induced inflammation and pulmonary fibrosis via inhibiting IL-6/STAT3 signaling pathway. J Ethnopharmacol. 2022; 283:114701.
[35] PULIVENDALA G, BALE S, GODUGU C. Honokiol: A polyphenol neolignan ameliorates pulmonary fibrosis by inhibiting TGF-β/Smad signaling, matrix proteins and IL-6/CD44/STAT3 axis both in vitro and in vivo. Toxicol Appl Pharmacol. 2020;391: 114913.
[36] 陈乾,付琦,王天明,等.胃癌中COL1A2表达及其与免疫细胞浸润关系的研究[J].医学理论与实践, 2023,36(12):1997-2000.
[37] 姚鸿飞.血清尿酸可通过抑制单核-巨噬细胞COL1A2和ITGA5表达减缓肺纤维化的形成[D].百色:右江民族医学院,2024.
[38] SONG C, LIU X, TAN W, et al. Fluorofenidone attenuates pulmonary inflammation and fibrosis by inhibiting the IL-11/MEK/ERK signaling pathway. Shock. 2022;58(2):137-146. |