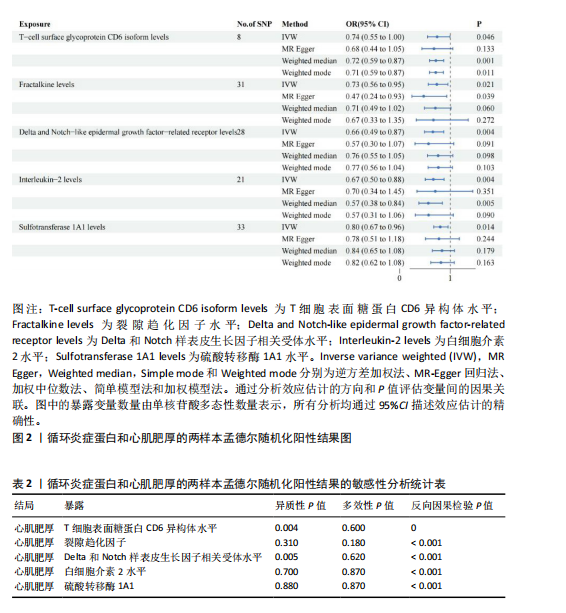
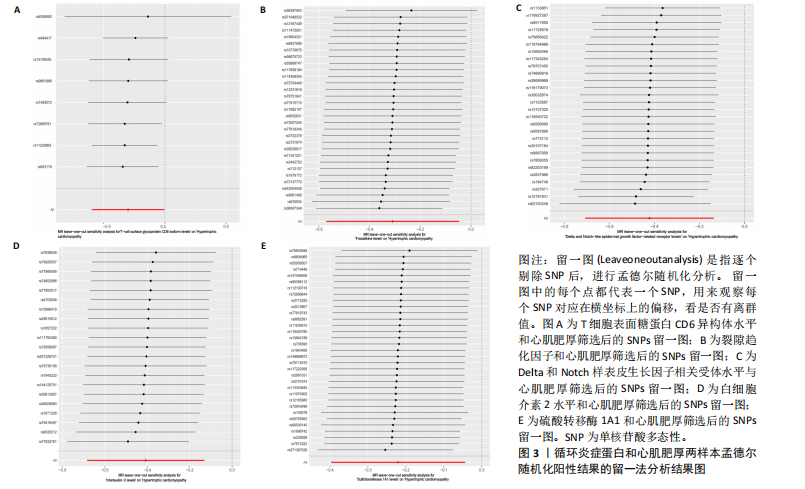
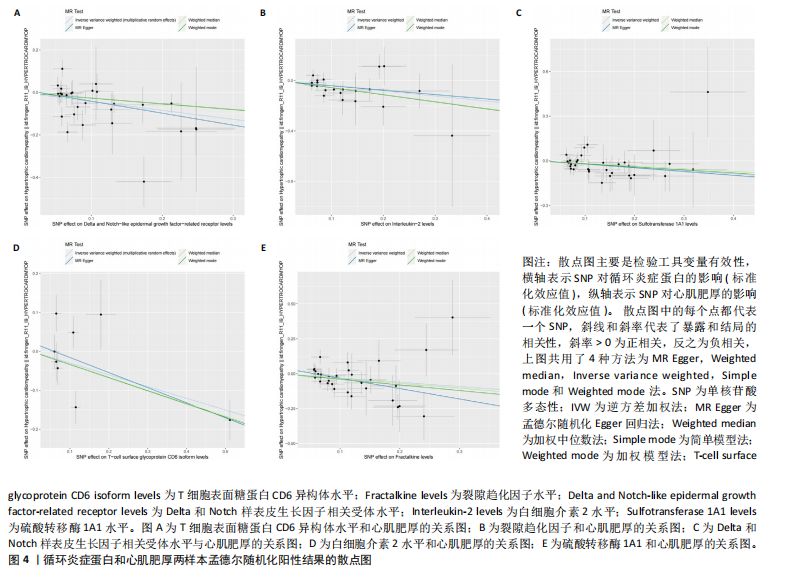
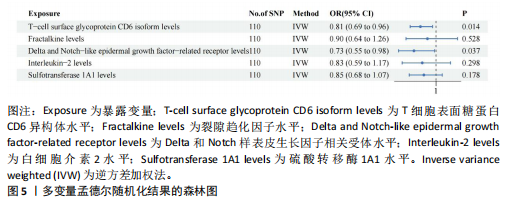
[1] D’AMATO G, LUXÁN G, DE LA POMPA JL. Notch signalling in ventricular chamber development and cardiomyopathy. FEBS J. 2016;283(23):4223-4237.
[2] FONSECA FA, IZAR MC. Role of inflammation in cardiac remodeling after acute myocardial infarction. Front Physiol. 2022;13:927163.
[3] DUBOIS-DERUY E, PEUGNET V, TURKIEH A, et al. Oxidative stress in cardiovascular diseases. Antioxidants (Basel). 2020;9(9): 864-864.
[4] NAUSHAD S, GAUCHER J, MEZDARI Z, et al. Chronic intermittent hypoxia triggers cardiac fibrosis: role of epididymal white adipose tissue senescent remodeling? Acta Physiol(Oxf). 2024;240(11):e14231.
[5] SHIMADA YJ, RAITA Y, LIANG LW, et al. Comprehensive proteomics profiling reveals circulating biomarkers of hypertrophic cardiomyopathy. Circ Heart Fail. 2021;14(7): e007849.
[6] DINA RADENKOVIĆ, SAURABH CHAWLA, GIANNI BOTTA, et al. Advanced cardiometabolic & inflammatory markers for prediction of cardiovascular disease and cancer. Eur Heart J. 2020. doi:10.1093/ehjci/ehaa946.2921.
[7] ARNOLD N, BLAUM C, GOßLING A, et al. C-reactive protein modifies lipoprotein(a)-related risk for coronary heart disease: the BiomarCaRE project. Eur Heart J. 2024; 45(12):1043-1054.
[8] MORITA H, REHM HL, MENESSES A, et al. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358(18):1899-1908.
[9] JI M, LIU Y, ZUO Z, et al. Downregulation of amphiregulin improves cardiac hypertrophy via attenuating oxidative stress and apoptosis. Biol Direct. 2022;17(1):21.
[10] BAZGIR F, NAU J, NAKHAEI‐RAD S, et al. The microenvironment of the pathogenesis of cardiac hypertrophy. Cells. 2023;12(13):1780-1780.
[11] MARON BJ, OMMEN SR, SEMSARIAN C, et al. Hypertrophic cardiomyopathy: present and future, with translation into contemporary cardiovascular medicine. J Am Coll Cardiol. 2014;64(1):83-99.
[12] ELLIOTT PM, ANASTASAKIS A, BORGER MA, et al. 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the task force for the diagnosis and management of hypertrophic cardiomyopathy of the european society of cardiology (ESC). Eur Heart J. 2014;35(39):2733-2779.
[13] CHENG Z, FANG T, HUANG J, et al. Hypertrophic cardiomyopathy: from phenotype and pathogenesis to treatment. Front Cardiovasc Med. 2021;8:722340.
[14] LOPES LR, AUNG N, VAN DUIJVENBODEN S, et al. Prevalence of hypertrophic cardiomyopathy in the UK Biobank Population. JAMA Cardiol. 2021;6(7):852-854.
[15] DAVEY SMITH G, HEMANI G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89-R98.
[16] WANG Y, SHEN H. Challenges and factors that influencing causal inference and interpretation, based on Mendelian randomization studies. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(8):1231-1236.
[17] GUO Z, WANG Z, GAO Z, et al. The status and trends of mitochondrial dynamics research: a global bibliometric and visualized analysis. J Bioenerg Biomembr. 2023;55(1):43-57.
[18] ZHAO JH, STACEY D, ERIKSSON N, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol. 2023;24(9):1540-1551.
[19] LOUISE ACM, GEORGE DAVEY S, KATE T. The global randomization test: a Mendelian randomization falsification test for the exclusion restriction assumption. medRxiv (Cold Spring Harbor Laboratory). 2022. doi:10.1101/2022.05.03.22274459.
[20] BELL KJ, LOY C, CUST AE, et al. Mendelian randomization in cardiovascular research: establishing causality when there are unmeasured confounders. 2021;14(1): e005623.
[21] QINGYUAN Z, JINGSHU W, GIBRAN H, et al. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann Stat. 2020. doi:10.1214/19-aos1866.
[22] ZHIHAN X, ZICHEN W, TONGYU Z, et al. Bidirectional Mendelian randomization analysis of the genetic association between primary lung cancer and colorectal cancer. J Transl Med. 2023;21(1):722.
[23] STALEY JR, BLACKSHAW J, KAMAT MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207-3209.
[24] BURGESS S, BUTTERWORTH A, THOMPSON SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658-665.
[25] FANYE W, MINGZHE C, MINGHUI Z, et al. Causal association between oral disease and chronic obstructive pulmonary disease: a Mendelian Randomization study. Res Square. 2023. doi:10.21203/rs.3.rs-3179826/v1.
[26] BOWDEN J, DAVEY SMITH G, BURGESS S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525.
[27] BOWDEN J, DAVEY SMITH G, HAYCOCK PC, et al. Consistent estimation in Mendelian Randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016; 40(4):304-314.
[28] HARTWIG FP, DAVEY SMITH G, BOWDEN J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985-1998.
[29] ZHUANG X, YIN Q, YANG R, et al. Causal pathways in lymphoid leukemia: the gut microbiota, immune cells, and serum metabolites. Front Immunol. 2024;15: 1437869.
[30] EVANS DM, DAVEY SMITH G. Mendelian Randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet. 2015; 16(1):327-350.
[31] REBECCA CR, GEORGE DAVEY S. Commentary: Orienting causal relationships between two phenotypes using bidirectional Mendelian randomization. Int J Epidemiol. 2019;48(3):907-911
[32] COHEN JF, CHALUMEAU M, COHEN R, et al. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J Clin Epidemiol. 2015;68(3):299-306.
[33] ZHAO L, HU M, LI L. Identifying the genetic associations between diabetes mellitus and the risk of vitiligo. Clin Cosmet Investig Dermatol. 2024;17:2261-2271.
[34] BOWDEN J, HEMANI G, DAVEY SMITH G. Invited commentary: detecting individual and global horizontal pleiotropy in mendelian randomization-a job for the humble heterogeneity statistic? Am J Epidemiol. 2018;187(12):2681-2685.
[35] HEMANI G, TILLING K, DAVEY SMITH G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081.
[36] SLOB EAW, BURGESS S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44(4):313-329.
[37] FATIMA B, ASHISH P, DIPENDER G, et al. Disentangling the effects of traits with shared clustered genetic predictors using multivariable Mendelian randomization. Genet Epidemiol. 2022;46(7):415-429.
[38] ELEANOR S, TOM GR, TIM M, et al. Estimation of causal effects of a time-varying exposure at multiple time points through multivariable mendelian randomization. PLoS Genet. 2022;18(7):e1010290-e1010290.
[39] MARIE CS, CHIARA A, KAIDO L, et al. Quantifying the role of transcript levels in mediating DNA methylation effects on complex traits and diseases. Nat Commun. 2022;13(1):7559.
[40] STEPHEN B, SIMON G. THOMPSON. Multivariable Mendelian Randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251-260.
[41] ZUBER V, GRINBERG NF, GILL D, et al. Combining evidence from Mendelian randomization and colocalization: review and comparison of approaches. Am J Hum Genet. 2022;109(5):767-782.
[42] CLAUDIA G, DAMJAN V, SCHADT EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383.
[43] WEI‐MING S, XIAOJING G, MENG D, et al. Systematic druggable genome-wide Mendelian randomisation identifies therapeutic targets for Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2023;94(11):954-961.
[44] MARK IM, GONÇALO RA, LON RC, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356-369.
[45] MARTA C, MARIO M, FERNANDO A, et al. Relevance of cd6-mediated interactions in the regulation of peripheral T-cell responses and tolerance. Front Immunol. 2017;8:594.
[46] Breuning J, Brown MH. T cell costimulation by CD6 is dependent on bivalent binding of a gads/slp-76 complex. Mol Cell Biol. 2017;37(11):e00071.
[47] BUGHANI U, SAHA A, KURIAKOSE A, et al. T cell activation and differentiation is modulated by a CD6 domain 1 antibody Itolizumab. PLoS One. 2017;12(7): e0180088.
[48] DA GLÓRIA VG, MARTINS DE ARAÚJO M, MAFALDA SANTOS A, et al. T cell activation regulates CD6 alternative splicing by transcription dynamics and SRSF1. J Immunol. 2014;193(1):391-399.
[49] HENRIQUES SN, OLIVEIRA L, SANTOS RF, et al. CD6-mediated inhibition of T cell activation via modulation of Ras. Cell Commun Signal. 2022;20(1):184.
[50] SALA V, GALLO S, LEO C, et al. Signaling to cardiac hypertrophy: insights from human and mouse RASopathies. Mol Med. 2012;18:938-947.
[51] FOLEY CN, STALEY JR, BREEN PG, et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021; 12(1):764.
[52] CHEN X, YANG Y, SUN S, et al. CX3C chemokine: hallmarks of fibrosis and ageing. Pharmacol Res. 2024;208:107348.
[53] WANG W, HU YF, PANG M, et al. BMP and Notch signaling pathways differentially regulate cardiomyocyte proliferation during ventricle regeneration. Int J Biol Sci. 2021;17(9):2157-2166.
[54] ZHANG Y, DONG Y, XIONG Z, et al. Sirt6-mediated endothelial-to-mesenchymal transition contributes toward diabetic cardiomyopathy via the Notch1 signaling pathway. Diabetes Metab Syndr Obes. 2020;13:4801-4808.
[55] POL JG, CAUDANA P, PAILLET J, et al. Effects of interleukin-2 in immunostimulation and immunosuppression. J Exp Med. 2019.doi:10.1084/jem.20191247.
[56] XIAO J, YU K, LI M, et al. The IL-2/anti-IL-2 complex attenuates cardiac ischaemia-reperfusion injury through expansion of regulatory T cells. Cell Physiol Biochem. 2017;44(5):1810-1827.
[57] ZENG Z, YU K, CHEN L, et al. Interleukin-2/anti-interleukin-2 immune complex attenuates cardiac remodeling after myocardial infarction through expansion of regulatory T cells. J Immunol Res. 2016; 2016:8493767.
[58] SAK K, EVERAUS H. Sulfotransferase 1a1 as a biomarker for susceptibility to carcinogenesis: from molecular genetics to the role of dietary flavonoids. Curr Drug Metab. 2016;17(6):528-541.
[59] WATANABE K, ARUMUGAM S, SREEDHAR R, et al. Small interfering RNA therapy against carbohydrate sulfotransferase 15 inhibits cardiac remodeling in rats with dilated cardiomyopathy. Cell Signal. 2015;27(7):1517-1524.
[60] NEMET I, FUNABASHI M, LI XS, et al. Microbe-derived uremic solutes enhance thrombosis potential in the host. mBio. 2023;14(6):e0133123.
[61] LIM YJ, SIDOR NA, TONIAL NC, et al. Uremic toxins in the progression of chronic kidney disease and cardiovascular disease: mechanisms and therapeutic targets. Toxins (Basel). 2021;13(2):142.
[62] HIDEKI F, YURIKO Y, YUSUKE Y, et al. Anti‐oxidative effect of AST‐120 on kidney injury after myocardial infarction. Br J Pharmacol. 2016;173(8):1302-1313.
[63] XIAOJIAO Z, XIAO-WEI H, YANJUN J, et al. Impact of inflammation and anti-inflammatory modalities on diabetic cardiomyopathy healing: from fundamental research to therapy. Int Immunopharmacol. 2023;123:110747.
|