[1] PEIXOTO P, CARTRON PF, SERANDOUR AA, et al. From 1957 to Nowadays: A Brief History of Epigenetics. Int J Mol Sci. 2020;21(20): 7571.
[2] WANG K, LIU H, HU Q, et al. Epigenetic regulation of aging: implications for interventions of aging and diseases. Signal Transduct Target Ther. 2022;7(1):374.
[3] SCHIAFFINO S, REGGIANI C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447-1531.
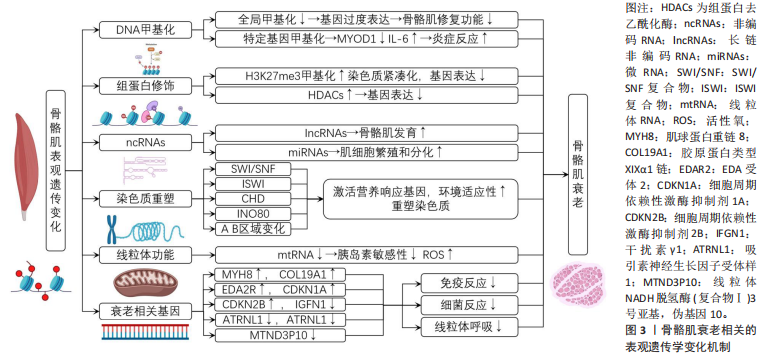
[4] 杨园,叶啟发,杨翼.运动调节衰老的表观遗传机制研究进展[J]. 中国运动医学杂志,2019,38(8):712-716.
[5] BARRèS R, ZIERATH JR. The role of diet and exercise in the transgenerational epigenetic landscape of T2DM. Nat Rev Endocrinol. 2016;12(8):441-451.
[6] 曹红, 吴迪, 魏瑜,等. 染色质结构蛋白变异与人类疾病[J]. 中国科学:生命科学,2023,53(11):1575-1594.
[7] TURNER DC, GORSKI PP, MAASAR MF, et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: the role of HOX genes and physical activity. Sci Rep. 2020;10(1):15360.
[8] 尹苗苗,牛燕媚,傅力.组蛋白去乙酰化酶在骨骼肌能量代谢调控中的潜在作用[J]. 中国糖尿病杂志,2013,21(2):189-192.
[9] MBADHI MN, TANG JM, ZHANG JX. Histone Lysine Methylation and Long Non-Coding RNA: The New Target Players in Skeletal Muscle Cell Regeneration. Front Cell Dev Biol. 2021;9:759237.
[10] 贾梅,浦敏铁.组蛋白修饰调控衰老过程的机制研究进展[J]. 中国科学:生命科学,2019,49(7):806-813.
[11] VOISIN S, HARVEY NR, HAUPT LM, et al. An epigenetic clock for human skeletal muscle. J Cachexia Sarcopenia Muscle. 2020;11(4):887-898.
[12] MASSENET J, GARDNER E, CHAZAUD B, et al. Epigenetic regulation of satellite cell fate during skeletal muscle regeneration. Skelet Muscle. 2021;11(1):4.
[13] PONSUKSILI S, TRAKOOLJUL N, BASAVARAJ S, et al. Epigenome-wide skeletal muscle DNA methylation profiles at the background of distinct metabolic types and ryanodine receptor variation in pigs. BMC Genomics. 2019;20(1):492.
[14] SAILANI MR, HALLING JF, MøLLER HD, et al. Lifelong physical activity is associated with promoter hypomethylation of genes involved in metabolism, myogenesis, contractile properties and oxidative stress resistance in aged human skeletal muscle. Sci Rep. 2019; 9(1):3272.
[15] WANG Y, YUAN Q, XIE L. Histone Modifications in Aging: The Underlying Mechanisms and Implications. Curr Stem Cell Res Ther. 2018;13(2): 125-135.
[16] 陈晓彤,赵文龙,孙林玉,等.非编码RNA来源的小肽:“微不足道”却“功能强大”[J]. 中山大学学报(自然科学版)(中英文), 2023,62(3):1-13.
[17] ZHAO Y, CHEN M, LIAN D, et al. Non-Coding RNA Regulates the Myogenesis of Skeletal Muscle Satellite Cells, Injury Repair and Diseases. Cells. 2019;8(9):988.
[18] 孔健达, 穆玉晶, 朱磊,等. 骨骼肌再生过程中卫星细胞调控机制及其生态位信号的作用[J]. 中国组织工程研究,2024,28(7): 1105-1111.
[19] LIU Q, DENG J, QIU Y, et al. Non-coding RNA basis of muscle atrophy. Mol Ther Nucleic Acids. 2021;26:1066-1078.
[20] LEE JH, KIM EW, CROTEAU DL, et al. Heterochromatin: an epigenetic point of view in aging. Exp Mol Med. 2020;52(9):1466-1474.
[21] BELL CG, LOWE R, ADAMS PD, et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 2019;20(1):249.
[22] GREVENDONK L, CONNELL NJ, MCCRUM C, et al. Impact of aging and exercise on skeletal muscle mitochondrial capacity, energy metabolism, and physical function. Nat Commun. 2021;12(1):4773.
[23] HEPPLE RT. Mitochondrial involvement and impact in aging skeletal muscle. Front Aging Neurosci. 2014;6:211.
[24] PEREZ K, CIOTLOS S, MCGIRR J, et al. Single nuclei profiling identifies cell specific markers of skeletal muscle aging, frailty, and senescence. Aging (Albany NY). 2022;14(23):9393-9422.
[25] TUMASIAN RA, 3RD, HARISH A, KUNDU G, et al. Skeletal muscle transcriptome in healthy aging. Nat Commun. 2021;12(1):2014.
[26] SHAVLAKADZE T, XIONG K, MISHRA S, et al. Age-related gene expression signatures from limb skeletal muscles and the diaphragm in mice and rats reveal common and species-specific changes. Skelet Muscle. 2023;13(1):11.
[27] YIN Y, MORGUNOVA E, JOLMA A, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356(6337):eaaj2239.
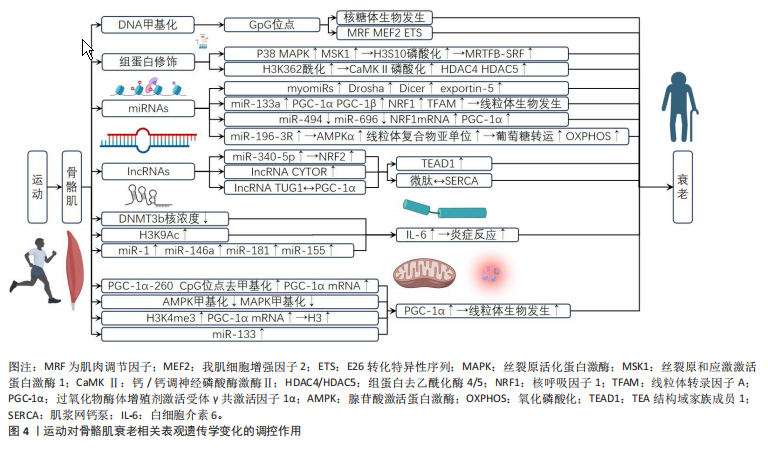
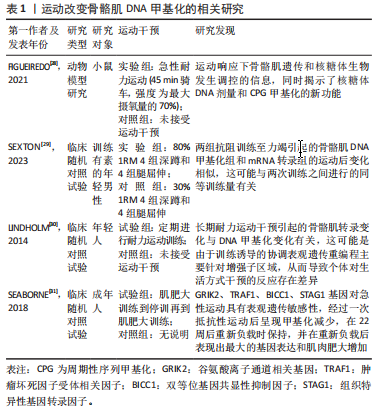
[28] FIGUEIREDO VC, WEN Y, ALKNER B, et al. Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. J Physiol. 2021;599(13):3363-3384.
[29] SEXTON CL, GODWIN JS, MCINTOSH MC, et al. Skeletal Muscle DNA Methylation and mRNA Responses to a Bout of Higher versus Lower Load Resistance Exercise in Previously Trained Men. Cells. 2023; 12(2):263.
[30] LINDHOLM ME, MARABITA F, GOMEZ-CABRERO D, et al. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics. 2014;9(12):1557-1569.
[31] SEABORNE RA, STRAUSS J, COCKS M, et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci Rep. 2018;8(1): 1898.
[32] SEABORNE RA, SHARPLES AP. The Interplay Between Exercise Metabolism, Epigenetics, and Skeletal Muscle Remodeling. Exerc Sport Sci Rev. 2020;48(4):188-200.
[33] ŚWIATOWY WJ, ZIELIŃSKI J, OSIELSKA MA, et al. No dynamic changes in the expression of genes related to the epigenetic mechanism during acute exercise. J Appl Genet. 2023;64(1):81-87.
[34] 张艺蓉,陈力方,王维蓉. HDACs去乙酰化酶非依赖性作用在疾病中的研究进展[J]. 生命科学,2023,35(5):583-591.
[35] NAGHAVI MOGHADAM AA, SHIRAVAND M, REZAPOUR S, et al. Effect of a session of intensive exercise with ginseng supplementation on histone H3 protein methylation of skeletal muscle of nonathlete men. Mol Genet Genomic Med. 2019;7(5):e651.
[36] SOLAGNA F, NOGARA L, DYAR KA, et al. Exercise-dependent increases in protein synthesis are accompanied by chromatin modifications and increased MRTF-SRF signalling. Acta Physiol (Oxf). 2020;230(1):e13496.
[37] JACQUES M, HIAM D, CRAIG J, et al. Epigenetic changes in healthy human skeletal muscle following exercise- a systematic review. Epigenetics. 2019;14(7):633-648.
[38] RUSSELL AP, LAMON S, BOON H, et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J Physiol. 2013;591(18):4637-4653.
[39] NIE Y, SATO Y, WANG C, et al. Impaired exercise tolerance, mitochondrial biogenesis, and muscle fiber maintenance in miR-133a-deficient mice. Faseb J. 2016;30(11):3745-3758.
[40] 陈圣菊,王飞,李文炯,等.小鼠运动敏感型血清miRNA的筛选[J]. 航天医学与医学工程,2018,31(2):224-228.
[41] 陈彩珍,李德深,邱守涛,等.肌组织特异性microRNA介导的骨骼肌衰减机制及运动的干预研究[J]. 西安体育学院学报,2017, 34(6):705-713.
[42] SUN Y, CUI D, ZHANG Z, et al. Voluntary wheel exercise alters the levels of miR-494 and miR-696 in the skeletal muscle of C57BL/6 mice. Comp Biochem Physiol B Biochem Mol Biol. 2016;202:16-22.
[43] MASSART J, SJöGREN RJO, EGAN B, et al. Endurance exercise training-responsive miR-19b-3p improves skeletal muscle glucose metabolism. Nat Commun. 2021;12(1):5948.
[44] HICKS SD, JACOB P, MIDDLETON FA, et al. Distance running alters peripheral microRNAs implicated in metabolism, fluid balance, and myosin regulation in a sex-specific manner. Physiol Genomics. 2018; 50(8):658-667.
[45] KOPP F, MENDELL JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172(3):393-407.
[46] THOMSON DW, DINGER ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272-283.
[47] WU YL, LIN ZJ, LI CC, et al. Epigenetic regulation in metabolic diseases: mechanisms and advances in clinical study. Signal Transduct Target Ther. 2023;8(1):98.
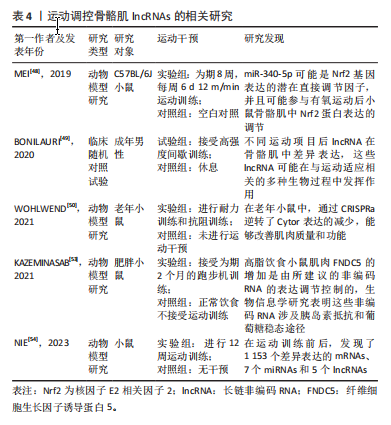
[48] MEI T, LIU Y, WANG J, et al. miR‑340‑5p: A potential direct regulator of Nrf2 expression in the post‑exercise skeletal muscle of mice. Mol Med Rep. 2019;19(2):1340-1348.
[49] BONILAURI B, DALLAGIOVANNA B. Long Non-coding RNAs Are Differentially Expressed After Different Exercise Training Programs. Front Physiol. 2020;11:567614.
[50] WOHLWEND M, LAURILA PP, WILLIAMS K, et al. The exercise-induced long noncoding RNA CYTOR promotes fast-twitch myogenesis in aging. Sci Transl Med. 2021;13(623):eabc7367.
[51] 陈雪飞,张靓. 肌浆网钙泵调节肽的研究进展[J]. 生命的化学, 2017,37(6):1005-1011.
[52] 潘剑锋,尚方正,马荣,等.长非编码RNA编码微肽的研究进展[J]. 生物工程学报,2022,38(9):3194-3214.
[53] KAZEMINASAB F, MARANDI SM, BAHARLOOIE M, et al. Aerobic exercise modulates noncoding RNA network upstream of FNDC5 in the Gastrocnemius muscle of high-fat-diet-induced obese mice. J Physiol Biochem. 2021;77(4):589-600.
[54] NIE M, LIU Q, YAN C. Construction of a novel lncRNA-miRNA-mRNA competing endogenous RNA network in muscle in response to exercise training. Gen Physiol Biophys. 2023;42(2):123-133.
[55] TREWIN AJ, SILVER J, DILLON HT, et al. Long non-coding RNA Tug1 modulates mitochondrial and myogenic responses to exercise in skeletal muscle. BMC Biol. 2022;20(1):164.
[56] HUNTER DJ, JAMES L, HUSSEY B, et al. Impact of aerobic exercise and fatty acid supplementation on global and gene-specific DNA methylation. Epigenetics. 2019;14(3):294-309.
[57] COCO M, PERCIAVALLE V, CAVALLARI P, et al. Effects of age and sex on epigenetic modification induced by an acute physical exercise. Medicine (Baltimore). 2017;96(44):e8325.
[58] SANDONà M, CAVIOLI G, RENZINI A, et al. Histone Deacetylases: Molecular Mechanisms and Therapeutic Implications for Muscular Dystrophies. Int J Mol Sci. 2023;24(5):4306.
[59] LI D, WANG P, WEI W, et al. Serum MicroRNA Expression Patterns in Subjects After the 5-km Exercise Are Strongly Associated With Cardiovascular Adaptation. Front Physiol. 2021;12:755656.
[60] YUAN X, BERG N, LEE JW, et al. MicroRNA miR-223 as regulator of innate immunity. J Leukoc Biol. 2018;104(3):515-524.
[61] DE GONZALO-CALVO D, DáVALOS A, MONTERO A, et al. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J Appl Physiol (1985). 2015;119(2):124-134.
[62] PEDERSEN BK, AKERSTRöM TC, NIELSEN AR, et al. Role of myokines in exercise and metabolism. J Appl Physiol (1985). 2007;103(3): 1093-1098.
[63] HULSTON CJ, VENABLES MC, MANN CH, et al. Training with low muscle glycogen enhances fat metabolism in well-trained cyclists. Med Sci Sports Exerc. 2010;42(11):2046-2055.
[64] PALMER MJ. Functional segregation of synaptic GABAA and GABAC receptors in goldfish bipolar cell terminals. J Physiol. 2006;577(Pt 1): 45-53.
[65] WINBANKS CE, WEEKS KL, THOMSON RE, et al. Follistatin-mediated skeletal muscle hypertrophy is regulated by Smad3 and mTOR independently of myostatin. J Cell Biol. 2012;197(7):997-1008.
[66] 李兆进, 郑鹏程, 孔健达,等. 基于不同组织和器官角度回顾PGC-1α在运动抗衰老中的作用[J]. 中国组织工程研究,2024,28(29): 4717-4725.
[67] BAJPEYI S, COVINGTON JD, TAYLOR EM, et al. Skeletal Muscle PGC1α -1 Nucleosome Position and -260 nt DNA Methylation Determine Exercise Response and Prevent Ectopic Lipid Accumulation in Men. Endocrinology. 2017;158(7):2190-2199.
[68] MAASAR MF, TURNER DC, GORSKI PP, et al. The Comparative Methylome and Transcriptome After Change of Direction Compared to Straight Line Running Exercise in Human Skeletal Muscle. Front Physiol. 2021;12:619447.
[69] LOCHMANN TL, THOMAS RR, BENNETT JP, JR, et al. Epigenetic Modifications of the PGC-1α Promoter during Exercise Induced Expression in Mice. PLoS One. 2015;10(6):e0129647.
[70] MASUZAWA R, KONNO R, OHSAWA I, et al. Muscle type-specific RNA polymerase II recruitment during PGC-1α gene transcription after acute exercise in adult rats. J Appl Physiol (1985). 2018;125(4):1238-1245. |