Chinese Journal of Tissue Engineering Research ›› 2017, Vol. 21 ›› Issue (20): 3208-3215.doi: 10.3969/j.issn.2095-4344.2017.20.016
Previous Articles Next Articles
Accurate determination of HLA ambiguous results based on group-specific haploid full-length sequencing
Wang Song-xing1, Yang Hui2, He Liu-mei1, Hong Wen-xu1, Zou Hong-yan1, Xu Yun-ping1
- 1Institute of Transfusion Medicine of Shenzhen Blood Center, Shenzhen 518035, Guangdong Povince, China; 2Shenzhen Jin Baihui Biological Co., Ltd., Shenzhen 518035, Guangdong Province, China
-
Revised:2017-05-18Online:2017-07-18Published:2017-07-28 -
Contact:Zou Hong-yan, Chief technician, Institute of Transfusion Medicine of Shenzhen Blood Center, Shenzhen 518035, Guangdong Povince, China; Xu Yun-ping, M.D., Associate chief technician, Institute of Transfusion Medicine of Shenzhen Blood Center, Shenzhen 518035, Guangdong Povince, China -
About author:Wang Song-xing, Technician-in-charge, Institute of Transfusion Medicine of Shenzhen Blood Center, Shenzhen 518035, Guangdong Povince, China -
Supported by:the Medical Research Foundation of Guangdong Province, No. A2016222; the Special Foundation for Strategic Emerging Industries Development in Shenzhen, No. JSGG20160328103642937; the Science and Technology Research and Development Foundation of Shenzhen, No. JCYJ20160427172335974; the Research Project of Shenzhen Health and Family Planning System, No. 201401077
CLC Number:
Cite this article
Wang Song-xing, Yang Hui, He Liu-mei, Hong Wen-xu, Zou Hong-yan, Xu Yun-ping. Accurate determination of HLA ambiguous results based on group-specific haploid full-length sequencing[J]. Chinese Journal of Tissue Engineering Research, 2017, 21(20): 3208-3215.
share this article

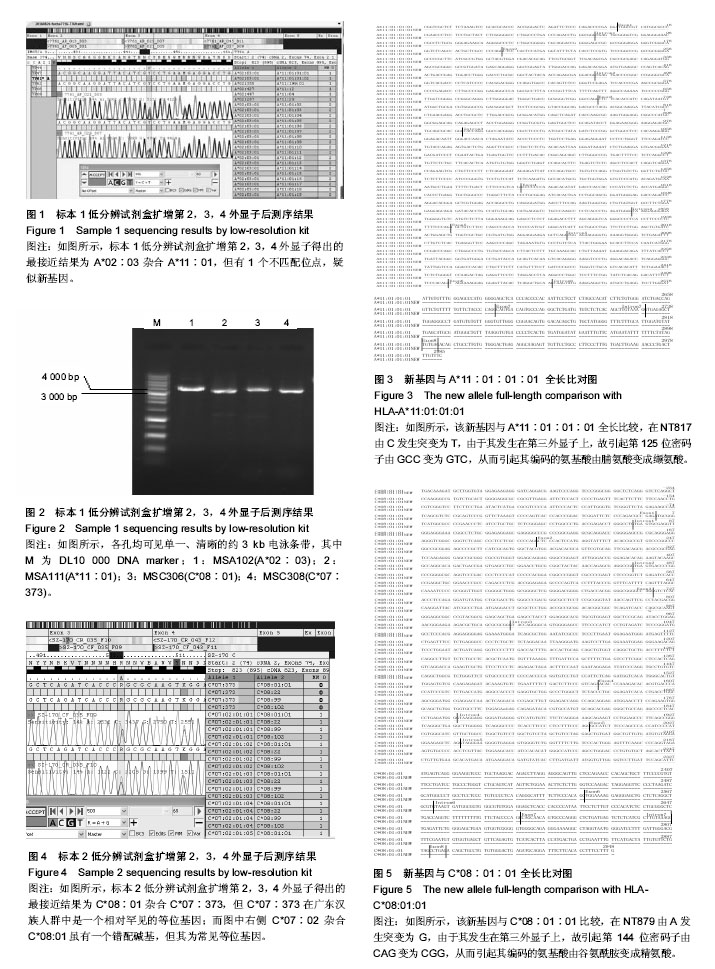
2.1 HLA组特异性单倍型全长测序分型方法解决A位点模棱两可结果 标本1低分辨试剂盒扩增第2,3,4外显子得出的结果为A*02∶03杂合A*11∶01(图1),但有1个不匹配位点,疑似新基因。根据HLA全长测序分型试剂盒(组特异性单倍型PCR-SBT法)A位点测序引物信息表(表1)用MSA102扩增A*02, MSA111扩增A*11,扩增产物电泳图分别如图2中第1、2泳道所示,然后进行测序分析得出分型结果为A*02∶03∶01杂合新等位基因,该基因与A*11∶01∶01∶01全长比较,在NT817由C发生突变为T(图3),由于其发生在第三外显子上,故引起第125位密码子由GCC变为GTC,从而引起其编码的氨基酸由脯氨酸变成缬氨酸。 2.2 HLA组特异性单倍型全长测序分型方法解决C位点模棱两可结果 标本2低分辨试剂盒扩增第2,3,4外显子得出的结果为C*08∶01杂合C*07∶373(图4),但C*07∶373在中国汉族人群中是一个相对罕见的等位基因;而图4中右侧黄色背景结果C*07∶02杂合C*08∶01为常见等位基因,虽有一个错配碱基,疑似新基因。根据HLA全长测序分型试剂盒(组特异性单倍型PCR-SBT法)C位点测序引物信息表(表2)用MSC306扩增C*08, MSC308扩增C*07,扩增产物电泳图分别如图2中第3、4泳道所示,然后进行测序分析得出分型结果为;C*07∶02∶01∶01杂合新等位基因,该基因与C*08∶01∶01比较,在NT879由A发生突变为G(图5),由于其发生在第三外显子上,故引起第144位密码子由CAG变为CGG,从而引起其编码的氨基酸由谷氨酰胺变成精氨酸。"
| [1] Jun Z,Brian D,Michael S, et al. Rapid Detection of Donor Cell Free DNA in Lung Transplant Recipients with Rejections Using Donor-recipient HLA Mismatch. Human Immunol. 2017; 78(4):342-349.[2] Jan S,Robert D,Anna K, et al. Role of HLA match on results of hematopoietic stem cell transplantations from unrelated donors in children with acute leukemia and bone marrow failure syndromes. Acta Haematologica Polonica. 2017;48(1): 48-53.[3] Paolo F,Linda C,Joseph T, et al. Providing Better-Matched Donors for HLA Mismatched Compatible Pairs Through Kidney Paired Donation. Transplantation. 2017;101(3): 642-648.[4] Katharina F,Dietrich W. HLA mismatching as a strategy to reduce relapse after alternative donor transplantation. Semin Hematol. 2016;53(2):57-64.[5] Varyani UT, Kute VB, Patel HV, et al. Participation of compatible donor to improve HLA matching can increase kidney transplant rate of O blood group patients. Clin Quer Nephrol. 2015;4(3-4):38-40.[6] Pauni? V, Gragert L, Schneider J, et al. Charting improvements in US registry HLA typing ambiguity using a typing resolution score. Human immunology. 2016;77(7):542-549.[7] Wang D, He L, Zou H, et al. Analysis of ambiguities in HLA sequencing-based typing and its solutions. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2015;32(1): 44-48.[8] Jun J, Hwang K, Kim S, et al. Estimation of the 6‐digit level allele and haplotype frequencies of HLA‐A,‐B, and‐C in Koreans using ambiguity‐solving DNA typing. Tissue Antigens. 2014;84(3):277-284.[9] Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13): 4576-4583. [10] Petersdorf EW. Optimal HLA matching in hematopoietic cell transplantation. Curr Opin Immunol. 2008;20(5):588-593.[11] Crocchiolo R, Zino E, Vago L, et al. Nonpermissive HLA-DPB1 disparity is a significant independent risk factor for mortality after unrelated hematopoietic stem cell transplantation. Blood. 2009;114(7):1437-1444.[12] Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366-374.[13] Hollenbach JA, Madbouly A, Gragert L, et al. A combined DPA1-DPB1 amino acid epitope is the primary unit of selection on the HLA-DP heterodimer. Immunogenetics. 2012;64(8):559-569.[14] Spellman SR, Eapen M, Logan BR, et al. A perspective on the selection of unrelated donors and cord blood units for transplantation. Blood. 2012;120(2):259-265.[15] Lucan C, Pop L, Florian A, et al. HLA Genotyping using Next Generation Sequencing. Roman J Int Med. 2016,54(2): 98-104.[16] McGinnis MD, Conrad MP, Bouwens AG, et al. Automated, solid-phase sequencing of DRB region genes using T7 sequencing chemistry and dye-labeled primers. Tissue antigens. 1995;46(3 Pt 1)):173-179.[17] Versluis LF, Rozemuller E, Tonks S, et al. High-resolution HLA-DPB typing based upon computerized analysis of data obtained by fluorescent sequencing of the amplified polymorphic exon 2. Human Immunol. 1993;38(4):277-283.[18] Voorter CE, Rozemuller EH, de Bruyn-Geraets D, et al. Comparison of DRB sequence-based typing using different strategies. Tissue Antigens. 1997;49(5):471-476.[19] Scheltinga SA, Johnston-Dow LA, White CB, et al. A generic sequencing based typing approach for the identification of HLA-A diversity. Human Immunol. 1997;57(2):120-128.[20] Voorter CE, Kik MC, van den Berg-Loonen EM. High-resolution HLA typing for the DQB1 gene by sequence-based typing. Tissue Antigens. 1998;51(1):80-87.[21] Swelsen WT, Voorter CE, van den Berg-Loonen EM. Sequence analysis of exons 1, 2, 3, 4 and 5 of the HLA-B5/35 cross-reacting group. Tissue Antigens. 2002;60(3):224-234.[22] Swelsen WT, Voorter CE, van den Berg-Loonen EM. Ambiguities of human leukocyte antigen-B resolved by sequence-based typing of exons 1, 4, and 5. Tissue Antigens. 2004;63(3):248-254.[23] Robinson J, Halliwell JA, McWilliam H, et al. The IMGT/HLA database. Nucl Acids Res. 2013;41(Database issue): D1222-1227.[24] Mack SJ, Cano P, Hollenbach JA, et al. Common and well-documented HLA alleles: 2012 update to the CWD catalog. Tissue Antigens. 2013;81(4): 194-203.[25] Raphael C,Mirjana R,Seiamak B. Next-Generation Sequencing of the HLA locus: Methods and impacts on HLA typing, population genetics and disease association studies. Human Immunol. 2016;77(11):1016-1023.[26] Sharifeh K,Mansour S,Mahboobeh R, et al. Novel Multiplex Fluorescent PCR-Based Method for HLA Typing and Preimplantational Genetic Diagnosis of β-Thalassemia. Arch Med Res. 2016;47(4):293-298.[27] Eric T,Maureen M,Rosanne P, et al. Performance Characteristics and Validation of Next-Generation Sequencing for HLA typing. J Mol Diagn. 2016;18(5):668-675.[28] Moalic-Allain V,Mercier B,Gueguen P, et al. Next generation sequencing with a semi-conductor technology (Ion Torrent PGM™) for HLA typing: overall workflow performance and debate. Ann Biol Clin (Paris). 2016;74(4): 449-456.[29] Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens. 2012;80(1):1-11.[30] De Santis D, Dinauer D, Duke J, et al. 16(th) IHIW : review of HLA typing by NGS. Int J Immunogenet. 2013;40(1):72-76.[31] Smith LK. HLA typing by direct DNA sequencing. Methods in molecular biology. 2012;882:67-86.[32] Bentley G, Higuchi R, Hoglund B, et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue antigens. 2009;74(5):393-403.[33] Lind C, Ferriola D, Mackiewicz K, et al. Next-generation sequencing: the solution for high-resolution, unambiguous human leukocyte antigen typing. Human Immunol. 2010; 71(10):1033-1042.[34] Leenam D,Sunil P. Study of HLA allele frequency in Patel sub-population from India: Marrow Donor Registry India data. Indian J Transplant. 2016;10(3):73-74.[35] González-Galarza FF, Takeshita LY, Santos EJ, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(Database issue):D784-788.[36] Nagy M, Entz P, Otremba P, et al. Haplotype-specific extraction: auniversal method toresolve ambiguous genotypes and detect new alleles-demonstrated on HLA-B. Tissue Antigens. 2007;69:176-180.[37] Voorter CE, Palusci F, Tilanus MG. Sequence-based typing of HLA: an improved group-specific full-length gene sequencing approach. Methods Mol Biol. 2014;1109:101-114.[38] Profaizer T,Lázár-Molnár E,Close D, et al. HLA genotyping in the clinical laboratory: comparison of next-generation sequencing methods. HLA. 2016;88(1-2):14-24.[39] Dalva K,Beksac M. Sequence-specific primed PCR (PCR-SSP) typing of HLA Class I and Class II alleles. Methods Mol Med. 2007;134: 51-60.[40] Saunders PM,Pymm P,Pietra G, et al. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med. 2016;213(5):791-807.[41] Shiina T. Next generation sequencing based HLA genomic and polymorphism analyses. MHC. 2015;22(2):84-94.[42] Assia G,Rachida R,Habiba A, et al. HLA Polymorphism in Algerian Children With Lymphomas. J Pediatr Hematol Oncol. 2015;37(8):458-461. |
| [1] | Yao Xiaoling, Peng Jiancheng, Xu Yuerong, Yang Zhidong, Zhang Shuncong. Variable-angle zero-notch anterior interbody fusion system in the treatment of cervical spondylotic myelopathy: 30-month follow-up [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1377-1382. |
| [2] | Jin Tao, Liu Lin, Zhu Xiaoyan, Shi Yucong, Niu Jianxiong, Zhang Tongtong, Wu Shujin, Yang Qingshan. Osteoarthritis and mitochondrial abnormalities [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1452-1458. |
| [3] | Wang Jing, Xiong Shan, Cao Jin, Feng Linwei, Wang Xin. Role and mechanism of interleukin-3 in bone metabolism [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(8): 1260-1265. |
| [4] | Hou Jingying, Guo Tianzhu, Yu Menglei, Long Huibao, Wu Hao. Hypoxia preconditioning targets and downregulates miR-195 and promotes bone marrow mesenchymal stem cell survival and pro-angiogenic potential by activating MALAT1 [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1005-1011. |
| [5] | Luo Xiaoling, Zhang Li, Yang Maohua, Xu Jie, Xu Xiaomei. Effect of naringenin on osteogenic differentiation of human periodontal ligament stem cells [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1051-1056. |
| [6] | Zhang Jinglin, Leng Min, Zhu Boheng, Wang Hong. Mechanism and application of stem cell-derived exosomes in promoting diabetic wound healing [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1113-1118. |
| [7] | An Weizheng, He Xiao, Ren Shuai, Liu Jianyu. Potential of muscle-derived stem cells in peripheral nerve regeneration [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1130-1136. |
| [8] | Li Jiajun, Xia Tian, Liu Jiamin, Chen Feng, Chen Haote, Zhuo Yinghong, Wu Weifeng. Molecular mechanism by which icariin regulates osteogenic signaling pathways in the treatment of steroid-induced avascular necrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 780-785. |
| [9] | Tang Jiping, Zhang Yeting. Exercise regulates adult hippocampal neurogenesis in Alzheimer’s disease: mechanism and role [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 798-803. |
| [10] | Chen Xiaoxu, Luo Yaxin, Bi Haoran, Yang Kun. Preparation and application of acellular scaffold in tissue engineering and regenerative medicine [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 591-596. |
| [11] | Kang Kunlong, Wang Xintao. Research hotspot of biological scaffold materials promoting osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 597-603. |
| [12] | Shen Jiahua, Fu Yong. Application of graphene-based nanomaterials in stem cells [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 604-609. |
| [13] | Zhang Tong, Cai Jinchi, Yuan Zhifa, Zhao Haiyan, Han Xingwen, Wang Wenji. Hyaluronic acid-based composite hydrogel in cartilage injury caused by osteoarthritis: application and mechanism [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 617-625. |
| [14] | Li Hui, Chen Lianglong. Application and characteristics of bone graft materials in the treatment of spinal tuberculosis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 626-630. |
| [15] | Gao Cangjian, Yang Zhen, Liu Shuyun, Li Hao, Fu Liwei, Zhao Tianyuan, Chen Wei, Liao Zhiyao, Li Pinxue, Sui Xiang, Guo Quanyi. Electrospinning for rotator cuff repair [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 637-642. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||