Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (10): 2662-2670.doi: 10.12307/2026.606
Previous Articles Next Articles
Cathepsin F as a potential serum biomarker for stroke risk prediction: GWAS database data analysis
Tian Meng1, 2, Lou Tianwei3, Zhang Yongchen1, Jia Hongling4
- 1Shandong University of Traditional Chinese Medicine, Jinan 250355, Shandong Province, China; 2Shandong Institute of Sport Science, Jinan 250102, Shandong Province, China; 3The Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan 250011, Shandong Province, China; 4The Second Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan 250001, Shandong Province, China
-
Received:2025-04-24Accepted:2025-05-20Online:2026-04-08Published:2025-09-01 -
Contact:Jia Hongling, MD, Chief physician, Doctoral supervisor, The Second Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan 250001, Shandong Province, China -
About author:Tian Meng, PhD, Shandong University of Traditional Chinese Medicine, Jinan 250355, Shandong Province, China; Shandong Institute of Sport Science, Jinan 250102, Shandong Province, China Lou Tianwei, MS, Physician, The Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan 250011, Shandong Province, China Tian Meng and Lou Tianwei contributed equally to this work. -
Supported by:Shandong Province Chinese Medicine Science and Technology Project, No. Z202250 (to JHL); Qilu Medical School Chinese Medicine Inheritance Project, No. 20229324 (to JHL [project participant]); Scientific Research Project of Shandong Institute of Sport Science, No. KYZX202411 (to TM)
CLC Number:
Cite this article
Tian Meng, Lou Tianwei, Zhang Yongchen, Jia Hongling . Cathepsin F as a potential serum biomarker for stroke risk prediction: GWAS database data analysis[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(10): 2662-2670.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
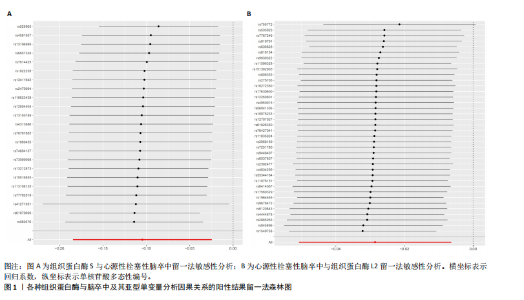
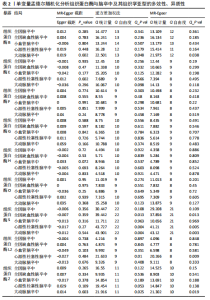
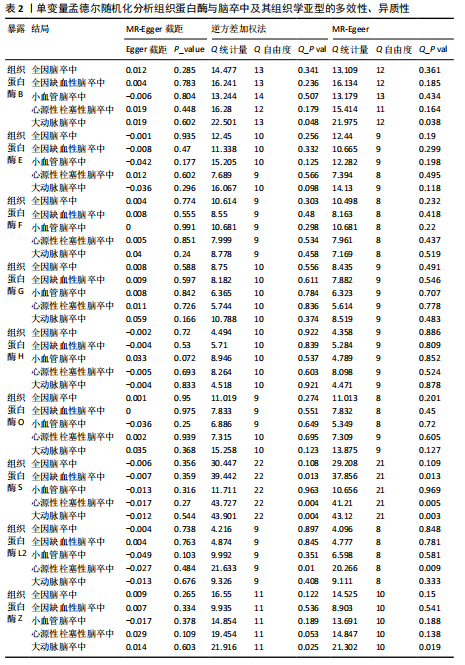
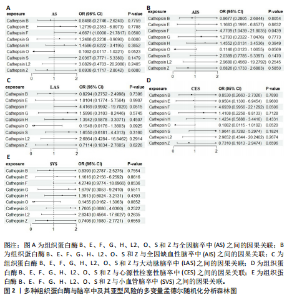
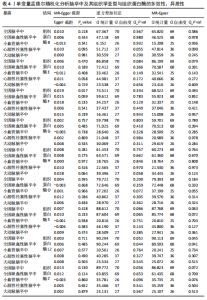
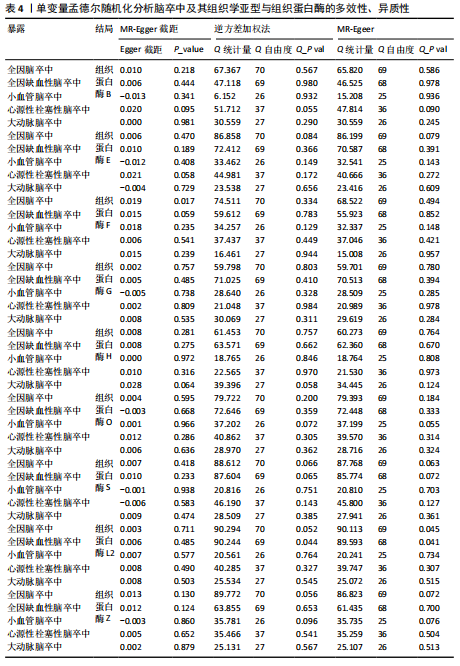
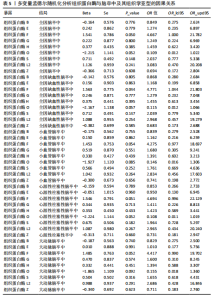
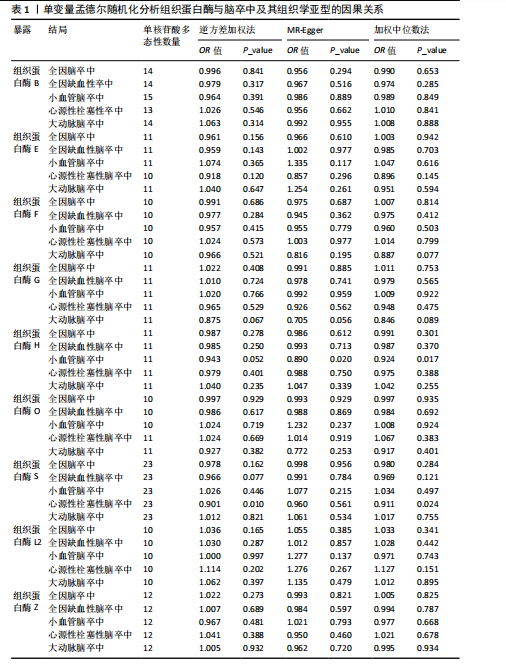
2.1 正向孟德尔随机化分析结果 为了评估各种组织蛋白酶对脑卒中及其亚型风险的影响,首先针对9种组织蛋白酶(组织蛋白酶B、E、F、G、H、L2、O、S和Z)和不同亚型脑卒中风险进行两样本孟德尔随机化分析。逆方差加权分析表明,较高的组织蛋白酶S水平对心源性栓塞性脑卒中的发生有抑制作用(OR=0.901,95%CI:0.832-0.976,P=0.010),见表1。尽管孟德尔随机化-逆方差加权法检测到Q_P val < 0.05,见表1,表明分析中存在异质性,但由于使用的是随机效应模型(逆方差加权法),这些异质性对分析结果的影响较小。另外,加权中位数法也支持上述相关性(OR=0.911,95%CI:0.840-0.988,P=0.024)。因此,认为较高的组织蛋白酶S水平与较低的心源性栓塞性脑卒中风险相关。同时,MR-Egger截距检验的P=0.270,验证了结果的稳健性,见表2。在留一法分析中,未发现单个单核苷酸多态性显著影响组织蛋白酶S对心源性栓塞性脑卒中的总体效应(图1A)。"
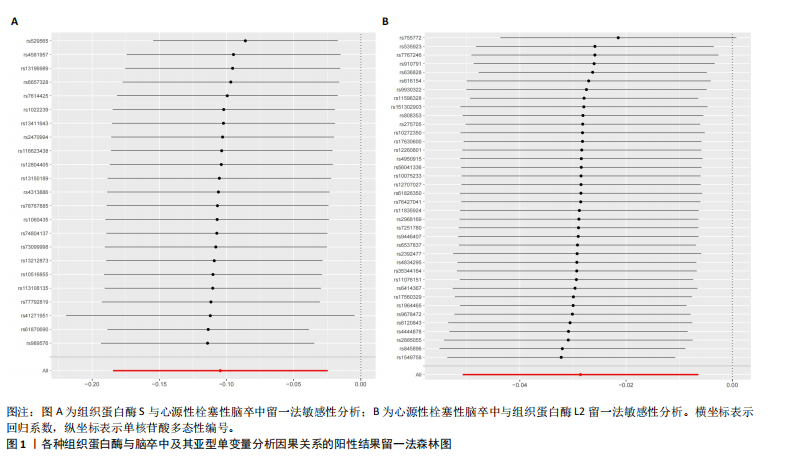
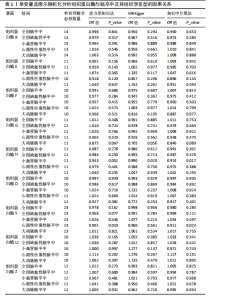
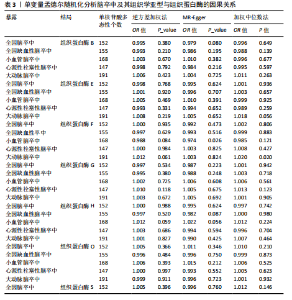
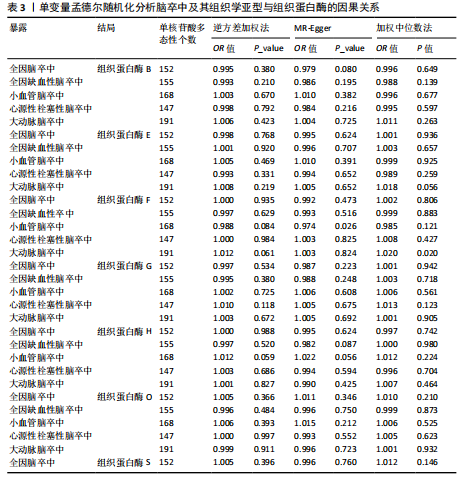
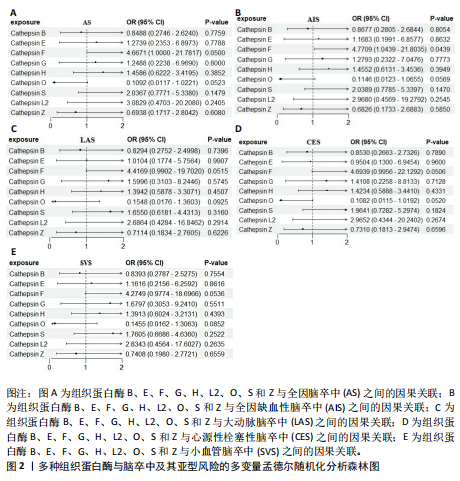
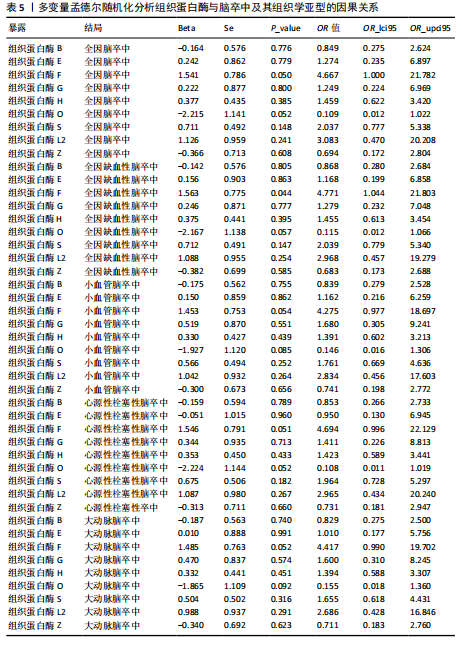
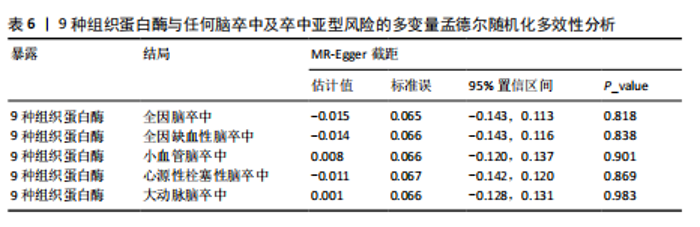
2.2 反向孟德尔随机化分析结果 为了探索反向因果关系的可能性,进行了反向孟德尔随机化分析,结果显示组织蛋白酶S与心源性脑卒中风险之间缺乏反向因果关系(表3)。然而,反向孟德尔随机化-逆方差加权分析结果表明,心源性栓塞性脑卒中可能导致组织蛋白酶L2水平的下降(OR=0.984,95%CI:0.972-0.998,P=0.020)。孟德尔随机化-逆方差加权分析检测的Q_pval=0.327,MR-Egger截距检验P=0.490,异质性和敏感性分析均验证了上述结果的稳健性(P > 0.05),见表4。在留一法分析中,排除每个单核苷酸多态性后,剩余单核苷酸多态性的效应值基本围绕整体效应值分布,且置信区间较窄,表明孟德尔随机化结果对单个单核苷酸多态性的依赖性较低,结果较为稳健(图1B)。没有证据支持其他亚型脑卒中与各种类型的组织蛋白酶之间存在因果关系,见表3。 2.3 多变量孟德尔随机化分析结果 进行了多变量孟德尔随机化分析,以评估多种组织蛋白酶的遗传易感性与不同亚型脑卒中风险的关系,结果显示,组织蛋白酶F水平升高与全因脑卒中风险增加相关,高水平的组织蛋白酶F最高能增加21.782倍的全因脑卒中发病风险(OR=4.667,95%CI=1.000-21.782,P=0.050)。 MR-Egger截距检验未检测到单核苷酸多态性多效性(P=0.818),见图2A,表5,6。组织蛋白酶F水平升高会增加缺血性脑卒中的发病风险,当组织蛋白酶F水平升高时,缺血性脑卒中的发病风险增加了4.771倍(OR=4.771),这一结果具有统计学意义(P=0.044),表明结果不太可能是偶然发生的。可以以95%CI断言,组织蛋白酶F水平升高与缺血性脑卒中发病风险的真实关系至少是1.044倍,可能高达21.804倍(95%CI:1.044-21.804)。MR-Egger截距分析也未显示水平多效性(P=0.838),见图2B,表5,6。其他类型组织蛋白酶与任何脑卒中及其亚型之间均未显示出统计学意义上的因果关系,见图2,表5。"
| [1] GBD 2016 LIFETIME RISK OF STROKE COLLABORATORS, FEIGIN VL, NGUYEN G, et al. Global, Regional, and Country-Specific Lifetime Risks of Stroke, 1990 and 2016. N Engl J Med. 2018;379(25):2429-2437. [2] GBD 2016 STROKE COLLABORATORS. Global, regional, and national burden of stroke, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18(5):439-458. [3] LINDSAY MP, NORRVING B, SACCO RL, et al. World Stroke Organization (WSO): Global Stroke Fact Sheet 2019. Int J Stroke. 2019;14(8):806-817. [4] KAHL A, BLANCO I, JACKMAN K, et al. Publisher Correction: Cerebral ischemia induces the aggregation of proteins linked to neurodegenerative diseases. Sci Rep. 2018;8(1):6802. [5] GIFFARD RG, XU L, ZHAO H, et al. Chaperones, protein aggregation, and brain protection from hypoxic/ischemic injury. J Exp Biol. 2004;207(Pt 18):3213-3220. [6] LIU Y, XUE X, ZHANG H, et al. Neuronal-targeted TFEB rescues dysfunction of the autophagy-lysosomal pathway and alleviates ischemic injury in permanent cerebral ischemia. Autophagy. 2019;15(3):493-509. [7] REISER J, ADAIR B, REINHECKEL T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest. 2010;120(10):3421-3431. [8] PATEL S, HOMAEI A, EL-SEEDI HR, et al. Cathepsins: Proteases that are vital for survival but can also be fatal. Biomed Pharmacother. 2018;105:526-532. [9] HU K, GAIRE BP, SUBEDI L, et al. Interruption of Endolysosomal Trafficking After Focal Brain Ischemia. Front Mol Neurosci. 2021; 14:719100. [10] YUAN D, HU K, LOKE CM, et al. Interruption of endolysosomal trafficking leads to stroke brain injury. Exp Neurol. 2021;345:113827. [11] CARLONI S, BUONOCORE G, BALDUINI W. Protective role of autophagy in neonatal hypoxia-ischemia induced brain injury. Neurobiol Dis. 2008;32(3):329-339. [12] WEN YD, SHENG R, ZHANG LS, et al. Neuronal injury in rat model of permanent focal cerebral ischemia is associated with activation of autophagic and lysosomal pathways. Autophagy. 2008;4(6):762-769. [13] RAMI A, LANGHAGEN A, STEIGER S. Focal cerebral ischemia induces upregulation of Beclin 1 and autophagy-like cell death. Neurobiol Dis. 2008;29(1):132-141. [14] ZHANG X, YAN H, YUAN Y, et al. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013;9(9):1321-1333. [15] Nakanishi H. Microglial cathepsin B as a key driver of inflammatory brain diseases and brain aging. Neural Regen Res. 2020; 15(1):25-29. [16] BRENNAN P, HAINAUT P, BOFFETTA P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12(4):399-408. [17] JULIAN TH, BODDY S, ISLAM M, et al. A review of Mendelian randomization in amyotrophic lateral sclerosis. Brain. 2022;145(3):832-842. [18] SUN BB, MARANVILLE JC, PETERS JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73-79. [19] MALIK R, CHAUHAN G, TRAYLOR M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537. [20] LIN Z, DENG Y, PAN W. Combining the strengths of inverse-variance weighting and Egger regression in Mendelian randomization using a mixture of regressions model. PLoS Genet. 2021; 17(11):e1009922. [21] BOWDEN J, DAVEY SMITH G, HAYCOCK PC, et al. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304-314. [22] BOWDEN J, DAVEY SMITH G, BURGESS S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. [23] COHEN JF, CHALUMEAU M, COHEN R, et al. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J Clin Epidemiol. 2015;68(3):299-306. [24] Gala H, Tomlinson I. The use of Mendelian randomisation to identify causal cancer risk factors: promise and limitations. J Pathol. 2020;250(5):541-554. [25] HILKENS NA, CASOLLA B, LEUNG TW, et al. Stroke. Lancet. 2024;403(10446):2820-2836. [26] EKKER MS, VERHOEVEN JI, VAARTJES I, et al. Stroke incidence in young adults according to age, subtype, sex, and time trends. Neurology. 2019;92(21):e2444-e2454. [27] YIIN GS, HOWARD DP, PAUL NL, et al. Age-specific incidence, outcome, cost, and projected future burden of atrial fibrillation-related embolic vascular events: a population-based study. Circulation. 2014;130(15):1236-1244. [28] BOGIATZI C, HACKAM DG, MCLEOD AI, et al. Secular trends in ischemic stroke subtypes and stroke risk factors. Stroke. 2014;45(11):3208-3213. [29] FERNLUND E, GYLLENHAMMAR T, JABLONOWSKI R, et al. Serum Biomarkers of Myocardial Remodeling and Coronary Dysfunction in Early Stages of Hypertrophic Cardiomyopathy in the Young. Pediatr Cardiol. 2017;38(4):853-863. [30] CUVELLIEZ M, VANDEWALLE V, BRUNIN M, et al. Circulating proteomic signature of early death in heart failure patients with reduced ejection fraction. Sci Rep. 2019;9(1):19202. [31] STYPMANN J, GLÄSER K, ROTH W, et al. Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin L. Proc Natl Acad Sci U S A. 2002; 99(9):6234-6239. [32] TANG Q, CAI J, SHEN D, et al. Lysosomal cysteine peptidase cathepsin L protects against cardiac hypertrophy through blocking AKT/GSK3beta signaling. J Mol Med (Berl). 2009;87(3):249-260. [33] BEVAN S, TRAYLOR M, ADIB-SAMII P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43(12):3161-3167. [34] OLSON OC, JOYCE JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15(12):712-729. [35] LIN Z, ZHAO S, LI X, et al. Cathepsin B S-nitrosylation promotes ADAR1-mediated editing of its own mRNA transcript via an ADD1/MATR3 regulatory axis. Cell Res. 2023;33(7):546-561. [36] LI J, CHEN Z, KIM G, et al. Cathepsin W restrains peripheral regulatory T cells for mucosal immune quiescence. Sci Adv. 2023;9(28):eadf3924. [37] ARORA K, HERROON M, AL-AFYOUNI MH, et al. Catch and Release Photosensitizers: Combining Dual-Action Ruthenium Complexes with Protease Inactivation for Targeting Invasive Cancers. J Am Chem Soc. 2018;140(43):14367-14380. [38] FRUGÉ AD, SMITH KS, BAIL JR, et al. Biomarkers Associated With Tumor Ki67 and Cathepsin L Gene Expression in Prostate Cancer Patients Participating in a Presurgical Weight Loss Trial. Front Oncol. 2020;10:544201. [39] BARARIA D, HILDEBRAND JA, STOLZ S, et al.Cathepsin S Alterations Induce a Tumor-Promoting Immune Microenvironment in Follicular Lymphoma. Cell Rep. 2020; 31(5):107522. [40] SONG L, WANG X, CHENG W, et al. Expression signature, prognosis value and immune characteristics of cathepsin F in non-small cell lung cancer identified by bioinformatics assessment. BMC Pulm Med. 2021;21(1):420. [41] WEI S, LIU W, XU M, et al. Cathepsin F and Fibulin-1 as novel diagnostic biomarkers for brain metastasis of non-small cell lung cancer. Br J Cancer. 2022;126(12):1795-1805. [42] SANTAMARÍA I, VELASCO G, PENDÁS AM, et al. Molecular cloning and structural and functional characterization of human cathepsin F, a new cysteine proteinase of the papain family with a long propeptide domain. J Biol Chem. 1999;274(20): 13800-13809. [43] ZDRAVKOVA K, MIJANOVIC O, BRANKOVIC A, et al. Unveiling the Roles of Cysteine Proteinases F and W: From Structure to Pathological Implications and Therapeutic Targets. Cells. 2024;13(11):917. [44] HSU A, PODVIN S, HOOK V. Lysosomal Cathepsin Protease Gene Expression Profiles in the Human Brain During Normal Development. J Mol Neurosci. 2018;65(4):420-431. [45] TANG CH, LEE JW, GALVEZ MG, et al. Murine cathepsin F deficiency causes neuronal lipofuscinosis and late-onset neurological disease. Mol Cell Biol. 2006;26(6):2309-2316. [46] DALTON JP, ROBINSON MW, BRINDLEY PJ. Cathepsin// BARRETT AJ, RAWLINGS ND, WOESSNER JF, eds. Handbook of Proteolytic Enzymes. 3rd ed. Cambridge, MA: Academic Press; 2013. [47] FINN RD, BATEMAN A, CLEMENTS J, et al.Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue): D222-D230. [48] DI FABIO R, MORO F, PESTILLO L, et al. Pseudo-dominant inheritance of a novel CTSF mutation associated with type B Kufs disease. Neurology. 2014;83(19):1769-1770. |
| [1] | Wang Zheng, Cheng Ji, Yu Jinlong, Liu Wenhong, Wang Zhaohong, Zhou Luxing. Progress and future perspectives on the application of hydrogel materials in stroke therapy [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(8): 2081-2090. |
| [2] | Gao Feng, Zhang Jun, Yu Wenjun, Chanyu Yujing, Zhao Le, Hu Yuting, Wang Junhua, Liu Yongfu. Effects of wrist-hand orthosis on hand dysfunction in stroke patients: a meta-analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(8): 2124-2131. |
| [3] | Tao Daiju, Su Haiyu, Wang Yuqi, Shen Zhiqiang, He Bo . Construction and identification of stable PC12 cell lines with high/low expression of miR-122-5p [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1790-1799. |
| [4] | Wu Zhilin, , He Qin, Wang Pingxi, Shi Xian, Yuan Song, Zhang Jun, Wang Hao . DYRK2: a novel therapeutic target for rheumatoid arthritis combined with osteoporosis based on East Asian and European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1569-1579. |
| [5] | Liu Hongtao, Wu Xin, Jiang Xinyu, Sha Fei, An Qi, Li Gaobiao. Causal relationship between age-related macular degeneration and deep vein thrombosis: analysis based on genome-wide association study data [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1602-1608. |
| [6] | Guo Ying, Tian Feng, Wang Chunfang. Potential drug targets for the treatment of rheumatoid arthritis: large sample analysis from European databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1549-1557. |
| [7] | Gao Zengjie, , Pu Xiang, Li Lailai, Chai Yihui, Huang Hua, Qin Yu. Increased risk of osteoporotic pathological fractures associated with sterol esters: evidence from IEU-GWAS and FinnGen databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1302-1310. |
| [8] | Liu Fengzhi, Dong Yuna, Tian Wenyi, Wang Chunlei, Liang Xiaodong, Bao Lin. Gene-predicted associations between 731 immune cell phenotypes and rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1311-1319. |
| [9] | Zhang Cuicui, Chen Huanyu, Yu Qiao, Huang Yuxuan, Yao Gengzhen, Zou Xu. Relationship between plasma proteins and pulmonary arterial hypertension and potential therapeutic targets [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1331-1340. |
| [10] | Yang Yuanyuan, Zhou Shanshan, Cheng Xiaofei, Feng Luye, Tang Jiqin. Network meta-analysis of non-invasive brain stimulation in the treatment of lower limb motor dysfunction after stroke [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1008-1018. |
| [11] | Zeng Hao, Sun Pengcheng, Chai Yuan, Huang Yourong, Zhang Chi, Zhang Xiaoyun. Association between thyroid function and osteoporosis: genome-wide data analysis of European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1019-1027. |
| [12] | Rong Xiangbin, , Zheng Haibo, Mo Xueshen, Hou Kun, Zeng Ping, . Plasma metabolites, immune cells, and hip osteoarthritis: causal inference based on GWAS data from European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1028-1035. |
| [13] | He Qiwang, , , Chen Bo, Liang Fuchao, Kang Zewei, Zhou Yuan, Ji Anxu, Tang Xialin, . Relationship between Alzheimer’s disease and sarcopenia and body mass index: analysis of GWAS datasets for European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1036-1046. |
| [14] | Ding Yu, Chen Jingwen, Chen Xiuyan, Shi Huimin, Yang Yudie, Zhou Meiqi, Cui Shuai, . Circulating inflammatory proteins and myocardial hypertrophy: large sample analysis of European populations from GWAS Catalog and FinnGen databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1047-1057. |
| [15] | Yu Weijie, Cao Dongdong, Guo Tianci, Niu Puyu, Yang Jialin, Wang Simin, Liu Aifeng. Risk prediction models of recurrence after percutaneous endoscopic lumbar discectomy: a systematic review and meta-analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 749-759. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||