Chinese Journal of Tissue Engineering Research ›› 2024, Vol. 28 ›› Issue (2): 208-215.doi: 10.12307/2023.874
Previous Articles Next Articles
Molecular docking analysis of the anti-inflammatory mechanism of Cibotium barometz and Epimedium for rheumatoid arthritis: animal experiment validation
Ran Lei1, 2, Han Haihui1, 2, Xu Bo1, 2, Wang Jianye1, 2, Shen Jun2, Xiao Lianbo2, 3, Shi Qi3
- 1Shanghai Guanghua School of Clinical Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 201203, China; 2Department of Joint Surgery, Guanghua Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 200052, China; 3Institute of Integrated Traditional Chinese and Western Medicine for Arthritis, Shanghai Institute of Traditional Chinese Medicine, Shanghai 200052, China
-
Received:2022-12-05Accepted:2023-01-29Online:2024-01-18Published:2023-06-30 -
Contact:Xiao Lianbo, MD, Chief physician, Doctoral supervisor, Department of Joint Surgery, Guanghua Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 200052, China; Institute of Integrated Traditional Chinese and Western Medicine for Arthritis, Shanghai Institute of Traditional Chinese Medicine, Shanghai 200052, China -
About author:Ran Lei, MD candidate, Physician, Shanghai Guanghua School of Clinical Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 201203, China; Department of Joint Surgery, Guanghua Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 200052, China -
Supported by:Shanghai Key Specialty Construction Project, No. shslczdzk04801 (to XLB); Shanghai Natural Foundation General Projects, Nos. 22ZR1453100 (to XLB) and 22ZR1453000 (to SJ); Construction of Key Areas in Shanghai's Clinical Highlands for Chinese Medicine, No. ZY(2021-2023)-0201-06 (to XLB)
CLC Number:
Cite this article
Ran Lei, Han Haihui, Xu Bo, Wang Jianye, Shen Jun, Xiao Lianbo, Shi Qi. Molecular docking analysis of the anti-inflammatory mechanism of Cibotium barometz and Epimedium for rheumatoid arthritis: animal experiment validation[J]. Chinese Journal of Tissue Engineering Research, 2024, 28(2): 208-215.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

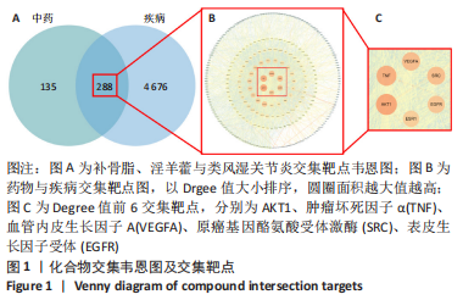
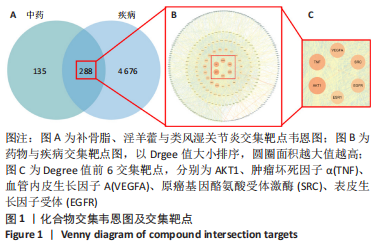
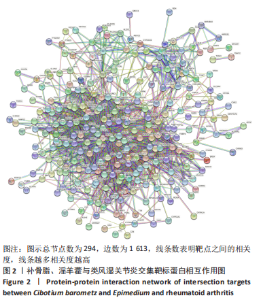
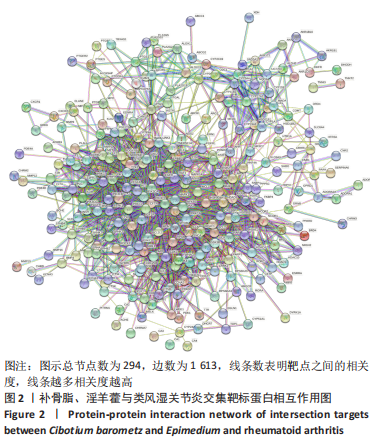
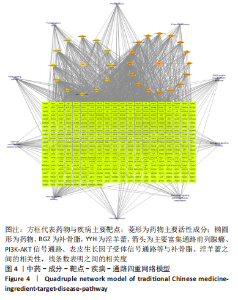
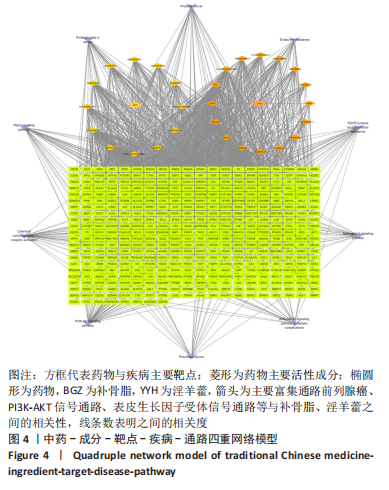
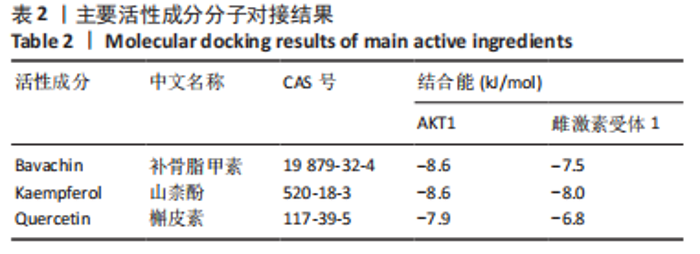
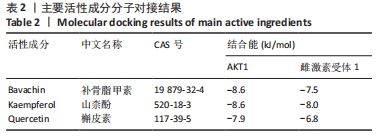
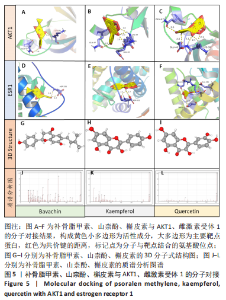
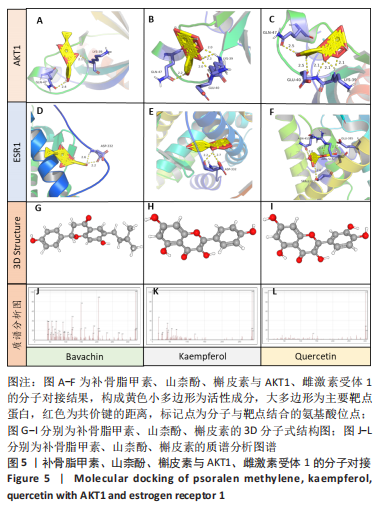
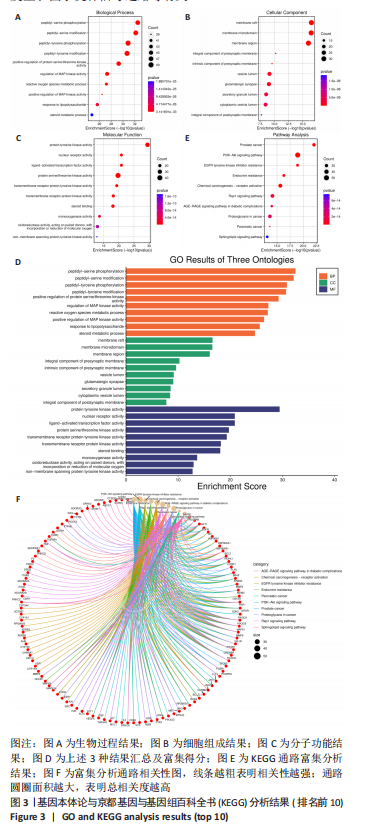
2.5 基因本体论与 KEGG富集分析 将2.3收集的交集靶点信息导入Metascape平台,以“H.Sapiens”提交,进行基因本体论和KEGG富集分析,获得生物过程2 232条,主要与丝氨酸蛋白磷酸化与修饰、丝氨酸/苏氨酸激酶正调控、活性氧代谢过程、MAP激酶活性正调节、脂多糖应答及类固醇的代谢过程等功能有关;细胞组成137条,主要与突触膜的成分和质膜形成等相关;分子功能获得273条,主要与丝氨酸激酶活性、丝氨酸/苏氨酸激酶活性、配体转录因子活性剂氧化还原酶活性相关,见图3。通过KEGG 通路富集分析获得202条相关信号通路,以P < 0.01、最小计数为3且富集因子> 1.5筛选,主要与前列腺癌、PI3K-AKT信号通路、表皮生长因子受体信号通路等有关。"
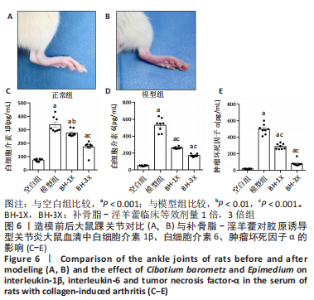
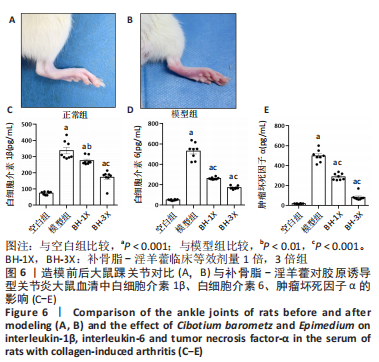
2.8 补骨脂-淫羊藿对CIA大鼠血清中白细胞介素1β、白细胞介素6、肿瘤坏死因子α的影响 造模前后大鼠踝关节对比见图6A,B。 白细胞介素1β:与空白组比较,其余3组较其表达均明显增加(P < 0.001);与模型组比较,BH-1X组表达降低(P < 0.01),BH-3X组表达降低(P < 0.001),见图6C。 白细胞介素6:与空白组比较,其余3组较其表达均明显增加(P < 0.001);与模型组比较,BH-1X组表达降低(P < 0.001),BH-3X组表达降低(P < 0.001),见图6D。 肿瘤坏死因子α:与空白组比较,其余3组较其表达均明显增加(P < 0.001);与模型组比较,BH-1X组表达降低(P < 0.001),BH-3X组表达降低(P < 0.001),见图6E。"
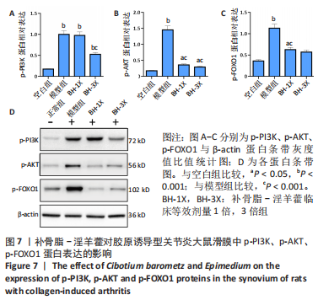
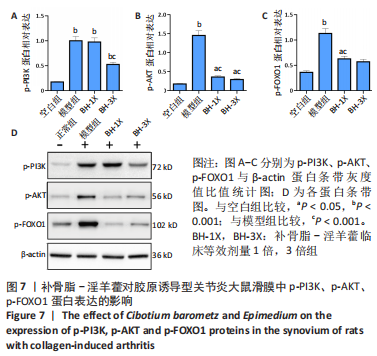
2.9 补骨脂-淫羊藿对胶原诱导型关节炎大鼠滑膜中p-PI3K、p-AKT、p-FOXO1蛋白表达的影响 见图7。 与空白组比较,其余3组p-PI3K表达均明显增加(P < 0.001);与模型组比较,BH-1X组表达无差异(P > 0.05),BH-3X组表达降低(P < 0.001),见图7A。 与空白组比较,模型组p-AKT表达明显增加(P < 0.001),BH-1X组、BH-3X组表达均增加(P < 0.05);与模型组比较,BH-1X组、BH-3X组表达均明显降低(P < 0.001),见图7B。 与空白组比较,模型组p-FOXO1表达明显增加(P < 0.001),BH-1X组、BH-3X组表达均增加(P < 0.05);与模型组比较,BH-1X组、BH-3X组表达均明显降低(P < 0.001),见图7C。 "
| [1] TIAN X, WANG Q, LI M, et al. 2018 Chinese guidelines for the diagnosis and treatment of rheumatoid arthritis. Rheumatol Immunol Res. 2021;2:1-14. [2] CHAURASIA N, SINGH A, SINGH IL, et al. Cognitive dysfunction in patients of rheumatoid arthritis. J Fam Med Prim Care. 2020;9:2219-2225. [3] LASSERE MN, RAPPO J, PORTEK IJ, et al. How many life years are lost in patients with rheumatoid arthritis? Secular cause-specific and all-cause mortality in rheumatoid arthritis, and their predictors in a long-term Australian cohort study. Intern Med J. 2013;43(1):66-72. [4] 童荣生.生物制剂治疗类风湿关节炎合理用药中国专家共识[J].中国新药杂志,2022,31(21):2174-2184. [5] 《类风湿关节炎超药品说明书用药中国专家共识》制定专家组.类风湿关节炎超药品说明书用药中国专家共识(2022版)[J].中华医学杂志,2022,102(15): 1076-1085. [6] 孙青,谢瑶,赵卫红,等.大剂量甲氨蝶呤治疗毒副作用系统分析[J].中国当代儿科杂志,2017,19(7):781-785. [7] SMOLEN JS, LANDEWÉ RBM, BIJLSMA JWJ, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis. 2020;79(6):685-699. [8] 顾娜沙. TNF-α抑制剂治疗类风湿关节炎患者用药现状的单中心观察研究[D].乌鲁木齐:新疆医科大学,2022. [9] 王慧,王永福.RA生物制剂治疗[J].中国免疫学杂志,2017,33(12):1911-1916. [10] 肖涟波,席智杰,程少丹,等.施杞从热毒痹论治急性期类风湿关节炎[J].上海中医药杂志,2017,51(12):1-4. [11] 尹梦碟. 邱联群教授辨治类风湿关节炎的用药规律与相关文献研究[D].广州:广州中医药大学,2021. [12] 杨静,梁江,唐东昕,等.国医大师刘尚义教授治疗类风湿关节炎的用药规律分析[J].风湿病与关节炎,2021,10(5):5-10. [13] 苏清君,李鹏,边朝辉,等.补骨脂素通过炎症和Ras/Raf1介导对类风湿关节炎大鼠的保护作用[J].世界中医药,2021,16(21):3180-3184+3190. [14] 吴志明. 淫羊藿苷通过调控miR-223-3p/NLRP3信号轴缓解类风湿性关节炎的发生[D].南昌:南昌大学,2021. [15] 吴志明,向艳茹,祝小波,等.淫羊藿苷治疗类风湿关节炎作用机制的研究进展[J].风湿病与关节炎,2022,11(7):71-74+80. [16] WONG BR, JOSIEN R, LEE SY, et al. TRANCE (tumor necrosis factor [TNF]-related activation-induced cytokine), a new TNF family member predominantly expressed in T cells, is a dendritic cell-specific survival factor. J Exp Med. 1997;186(12):2075-2080. [17] 鲁亚奇,张晓,王金金,等.补骨脂化学成分及药理作用研究进展[J].中国实验方剂学杂志,2019,25(3):180-189. [18] 曾华婷,郭健,陈彦.淫羊藿素药理作用及其新型给药系统的研究进展[J].中草药,2020,51(20):5372-5380. [19] 肖涟波. 自拟蠲痹补肾方对类风湿关节炎炎症及骨破坏的防治作用[Z]. 上海: 上海中医药大学附属光华医院,2016-02-01. [20] CONFORTI A, DI COLA I, PAVLYCH V, et al. Beyond the joints, the extra-articular manifestations in rheumatoid arthritis. Autoimmun Rev. 2021;20(2):102735. [21] CATRINA AI, YTTERBERG AJ, REYNISDOTTIR G, et al. Lungs, joints and immunity against citrullinated proteins in rheumatoid arthritis. Nat Rev Rheumatol. 2014; 10(11):645-653. [22] WELLS PM, WILLIAMS FMK, MATEY H, et al. RA and the microbiome: Do host genetic factors provide the link? J Autoimmun. 2019;99:104-115. [23] 宋在鑫,柳清泳,王晓军,等.补肾通络汤联合甲氨蝶呤对类风湿关节炎患者炎症细胞因子、骨代谢标志物及血管新生相关因子的影响[J].现代生物医学进展,2020,20(19):3774-3778+3783. [24] 苏清君,李鹏,边朝辉,等.补骨脂素通过炎症和Ras/Raf1介导对类风湿关节炎大鼠的保护作用[J].世界中医药,2021,16(21):3180-3184+3190. [25] 王义翠,彭慧霞,夏子岚,等.淫羊藿苷药理作用及应用研究进展[J].中华中医药学刊:1-13[2023-02-04]. [26] 邱国伟,肖涟波,施杞.蠲痹强骨方对类风湿关节炎炎症和骨破坏的作用及机制研究进展[J].中医药临床杂志,2021,33(5):992-999. [27] 殷方明,肖涟波,张昀,等.柚皮苷抑制胶原诱导小鼠关节炎症作用机制的实验研究[J].中国骨与关节损伤杂志,2016,31(3):285-288. [28] 王宇,蒋嘉明,孔思远,等.补骨脂ADME及其相关毒性的研究进展[J].世界科学技术-中医药现代化,2017,19(2):276-281. [29] BAIER A, MEINECKEL I, GAY S, et al. Apoptosis in rheumatoid arthritis. Curr Opin Rheumatol. 2003;15:274-279. [30] YAMANISHI Y, BOYLE DL, GREEN DR, et al. p53 tumor suppressor gene mutations in fibroblast-like synoviocytes from erosion synovium and non-erosion synovium in rheumatoid arthritis. Arthritis Res Ther. 2004;7:R12-R18. [31] 李生贵,潘正启,叶铄,等.circGFRA1靶向miR-642a-5p调控类风湿关节炎滑膜成纤维细胞增殖、迁移和侵袭的机制研究[J].中国细胞生物学学报,2022, 44(7):1276-1284. [32] 康静婷,纪超.加兰他敏调节IL-1β/IL-1RA比率改善炎症微环境的机制探讨[J].J Chin Pharm Sci. 2022,31(10):773-781. [33] 高晓珺,解骏,肖涟波,等.TNF-α拮抗剂对胶原诱导大鼠类风湿关节炎骨破坏治疗作用的实验研究[J].中国骨质疏松杂志,2013,19(5):458-464. [34] 贺龙刚,高奥,林晓辉,等.青藤碱联合TNF-α抑制剂对RA模型大鼠骨破坏及体外破骨细胞活化的的影响[J].时珍国医国药,2021,32(10):2349-2352. [35] 徐琼,张恒,王辉.基于EGFR介导的信号通路探讨中药活性成分抗肿瘤研究进展[J].中国实验方剂学杂志,2023,29(6):246-253. [36] WANG Y, LYU Z, QIN Y, et al. FOXO1 promotes tumor progression by increased M2 macrophage infiltration in esophageal squamous cell carcinoma. Theranostics. 2020;10(25):11535-11548. [37] 张小娜,杜小正,袁博,等.TRAF6泛素化调控功能与RA发病机制的研究进展[J].中国免疫学杂志:1-15[2022-12-03]. [38] PERLMAN H, GEORGANAS C, PAGLIARI LJ, et al. Bcl-2 expression in synovial fibroblasts is essential for maintaining mitochondrial homeostasis and cell viability. J Immunol. 2000;164(10):5227-5235. [39] YU DH, LIU L, LU F, et al. Effect of total saponin of dioscoreae nipponicae rhizoma on PI3K/Akt signal pathway by in rIL-1β induced fibroblast-like synoviocytes. Chin J Exp Trad Med Formaul. 2012;18:199-202. [40] PAN DM, WANG Q, CAI XD, et al. Duanteng Yimu Decoction induces apoptosis through inhibition of PI3K/Akt pathway in rheumatoid arthritis fibroblast-like synoviocytes. Chin J Trad Chin Med Pharm. 2018;33:2051-2055. [41] ZOU LH, HOU CF, CHEN ZH, et al. Effects of Shuangwuxuanbi granule on the apoptosis of rheumatoid arthritis fibroblast-like synoviocytes and PI3K/Akt signaling pathway. Med J Chin PLA. 2018;43:721-726. [42] LIU Y, JIANG SS, CHEN XY, et al. Effect of Hei Gu Teng Zhui Feng Huo Luo Capsule on PI3K/AKT/HIF-1α protein signaling pathway in rheumatoid arthritis rats. Chin J Immun. 2019;35:2206-2212. [43] 王曼姝,孙思彤,袁宇,等.功能代谢组学在中药毒性评价中的研究策略及其应用[J].中草药,2023,54(2):349-358. [44] 唐进法,牛璐,王晓艳,等.补骨脂醇提物中8种成分在正常和脂多糖模型大鼠体内的组织分布比较研究[J].中国中药杂志,2022,47(24):6763-6779. [45] 时潇丽,杨德超,徐光临,等.含淫羊藿、补骨脂中成药的用药特点分析[J].中成药,2022,44(8):2744-2749. [46] 颜冬梅,陈家辉,卢文滢,等.基于2020年版《中国药典》的含补骨脂成方制剂分析[J].亚太传统医药,2022,18(11):33-38. |
| [1] | Liu Baofang, Xu Bin, Chen Lei. Pueraria decoction in the treatment of osteoarthritis: network pharmacology analysis and animal model validation [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(2): 193-199. |
| [2] | Zuo Jun, Ma Shaolin. Mechanism of beta-sitosterol on hypertrophic scar fibroblasts: an analysis based on network pharmacology [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(2): 216-223. |
| [3] | Wang Qian, Lu Ziang, Li Lihe, Lyu Chaoliang, Wang Meng, Zhang Cunxin. Sinomenine effectively inhibits interleukin-1beta-induced apoptosis in nucleus pulposus cells [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(2): 224-230. |
| [4] | Chen Simin, Hu Yingjun, Yan Wenrui, Ji Le, Shao Mengli, Sun Ze, Zheng Hongxing, Qi Shanshan. Establishment and evaluation of a streptozotocin-induced diabetic encephalopathy rat model [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(2): 237-241. |
| [5] | Ai Fangfang, Xiao Hongyan, Wang Fang, Zhu Yongzhao, Ma Lijun. Reversal effect of Lycium barbarum polysaccharide in combination with oxaliplatin on drug resistance of colon cancer stem cells [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 74-79. |
| [6] | Yang Ting, Ding Zhibin, Jiang Nan, Han Hongxia, Hou Miaomiao, Ma Cungen, Song Lijuan, Li Xinyi. Astrocytes regulate glial scar formation in cerebral ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 131-138. |
| [7] | Ma Suilu, He Zhijun, Liu Tao, Li Yan, He Yuanxu, He Bo, Wang Weiwei, Wei Xiaotao. Traditional Chinese medicine monomer in the prevention and treatment of flap necrosis by regulating “autophagy” [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 153-158. |
| [8] | Zheng Rongjiong, Deng Zerun, Han Dan, Sun Lihua. Mechanism underlying rat hepatocyte apoptosis regulated by exosomes derived from bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 44-49. |
| [9] | Sun Jing, Liao Jian, Sun Jiangling, Cheng Ping, Feng Hongchao. Recombinant human growth hormone promotes osteogenic differentiation of human dental pulp stem cells [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 56-61. |
| [10] | Shen Feiyan, Yao Jixiang, Su Shanshan, Zhao Zhongmin, Tang Weidong. Knockdown of circRNA WD repeat containing protein 1 inhibits proliferation and induces apoptosis of chondrocytes in knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(在线): 1-6. |
| [11] | Fang Xingyan, Tian Zhenli, Zhao Zheyi, Wen Ping, Xie Tingting. Effects of sodium arsenite on human umbilical vein endothelial cell injury and sphingosine kinases 1/sphingosine 1-phosphate signaling axis [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(在线): 1-7. |
| [12] | He Xi, Wan Yu, Tang Yuting, Yang Anning, Wu Kai, Jiao Yun, Bai Zhigang, Jiang Yideng, Shen Jiangyong. Erastin inhibits proliferation of hypertrophic scar fibroblasts [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(在线): 1-. |
| [13] | Ruan Ling, Wang Guanghua, Wu Rongping, Jin Zhan, Lyu Zhenqing, Zhang Nan, Li Shoubang. Correlation between exercise intensity and lipid metabolism disorder and oxidative stress in a high-diet rat model [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(8): 1149-1155. |
| [14] | Nie Chenchen, Su Kaiqi, Gao Jing, Fan Yongfu, Ruan Xiaodi, Yuan Jie, Duan Zhaoyuan, Feng Xiaodong. The regulatory role of circular RNAs in cerebral ischemia-reperfusion injury [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(8): 1286-1291. |
| [15] | Liu Xiaolin, Mu Xinyue, Ma Ziyu, Liu Shutai, Wang Wenlong, Han Xiaoqian, Dong Zhiheng. Effect of hydrogel-loaded simvastatin microspheres on osteoblast proliferation and differentiation [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(7): 998-1003. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||