Chinese Journal of Tissue Engineering Research ›› 2023, Vol. 27 ›› Issue (14): 2182-2193.doi: 10.12307/2023.424
Previous Articles Next Articles
Identification of potential biomarkers of pigmented villonodular synovitis by transcriptome analysis
Liu Yuan1, Liang Xuezhen1, 2, Xu Bo1, 2, Liu Jinbao1, 2, Li Gang1, 2
- 1Shandong University of Traditional Chinese Medicine, Jinan 250014, Shandong Province, China; 2Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan 250014, Shandong Province, China
-
Received:2022-06-06Accepted:2022-07-12Online:2023-05-18Published:2022-09-30 -
Contact:Li Gang, MD, Professor, Chief physician, Doctoral supervisor, Shandong University of Traditional Chinese Medicine, Jinan 250014, Shandong Province, China; Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan 250014, Shandong Province, China -
About author:Liu Yuan, Master, Shandong University of Traditional Chinese Medicine, Jinan 250014, Shandong Province, China -
Supported by:the National Natural Science Foundation of China (General Program), No. 81774333 (to LG); Shandong Province Traditional Chinese Medicine Classic Prescription Cooperative Innovation and Opening Project, No. 2019KFY17 (to XB); Shandong Provincial Key R&D Program, No. 2016GSF202022 (to LG)
CLC Number:
Cite this article
Liu Yuan, Liang Xuezhen, Xu Bo, Liu Jinbao, Li Gang. Identification of potential biomarkers of pigmented villonodular synovitis by transcriptome analysis[J]. Chinese Journal of Tissue Engineering Research, 2023, 27(14): 2182-2193.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
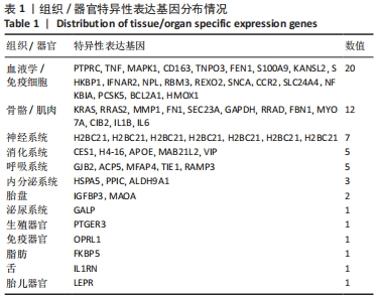
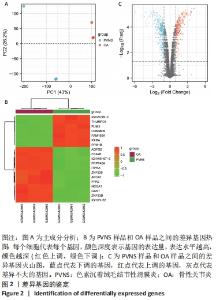
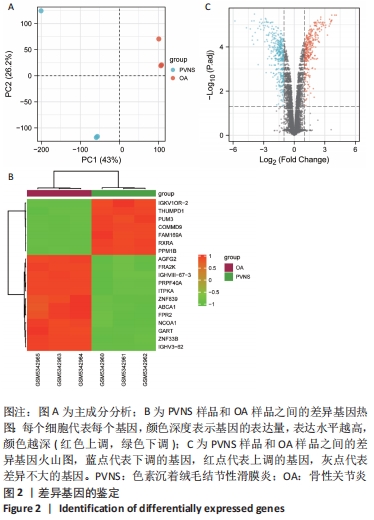
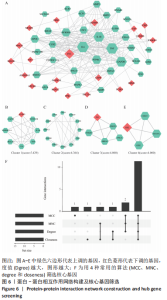
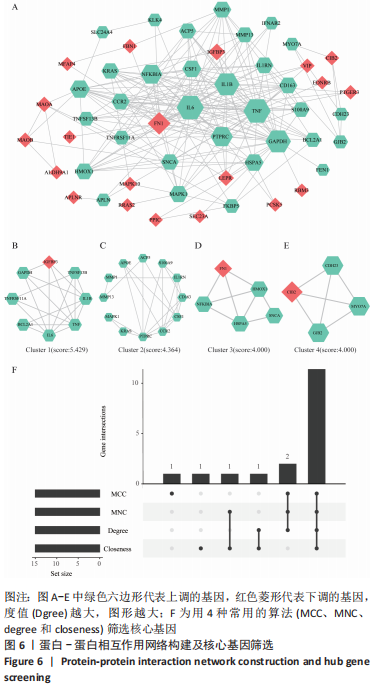
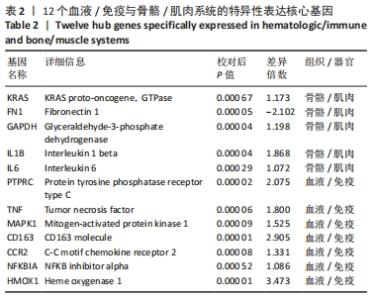
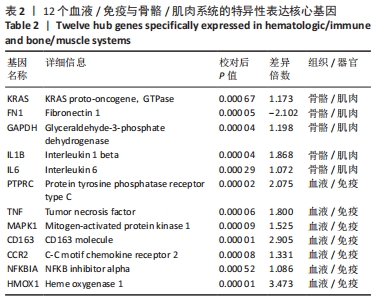
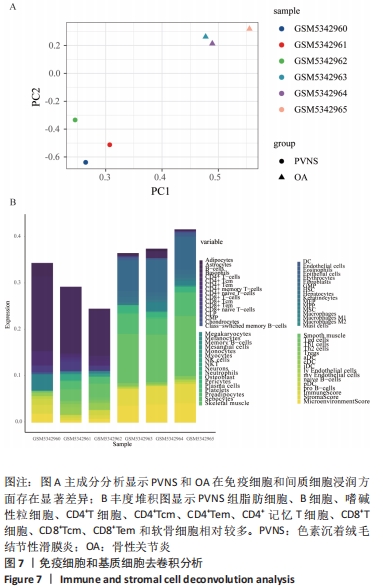
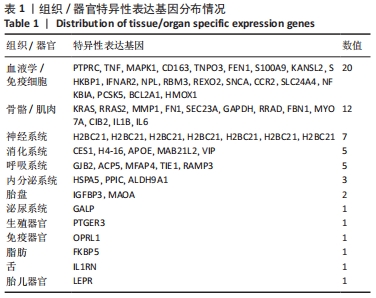
2.2 最终差异mRNA的获取 通过TTD、OMIM、GeneCard、DrugBank和DisGeNET数据库分别检索到与PVNS相关的基因为5,83,402,35和15个。去除重复之后共得到539个基因,与数据集得到的差异mRNA取交集后得到最终差异基因70个。 2.3 组织/器官特异性表达基因的鉴定 通过BioGPS共确定了60个组织/器官特异性表达基因,见表1。进一步分析发现大多数基因在血液/免疫系统中特异性表达,共有20个基因(20/60,33.33%);骨骼/肌肉系统共包含了12个特异性表达基因 (12/60,20.00%),其次为神经系统(7/60,11.67%)、呼吸系统(5/60,8.33%)、消化系统(5/60,8.33%)、内分泌系统(3/60,5.00%)、胎盘(2/60,3.33%);而生殖系统、泌尿系统、免疫器官、胎儿器官以及舌和脂肪组织所包括的特异性表达基因最少(1/60,1.67%)。"
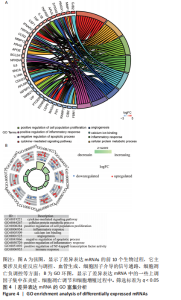
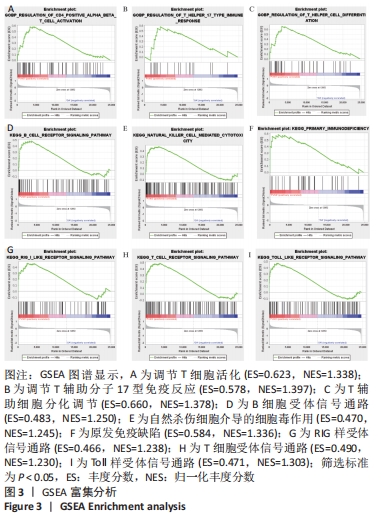
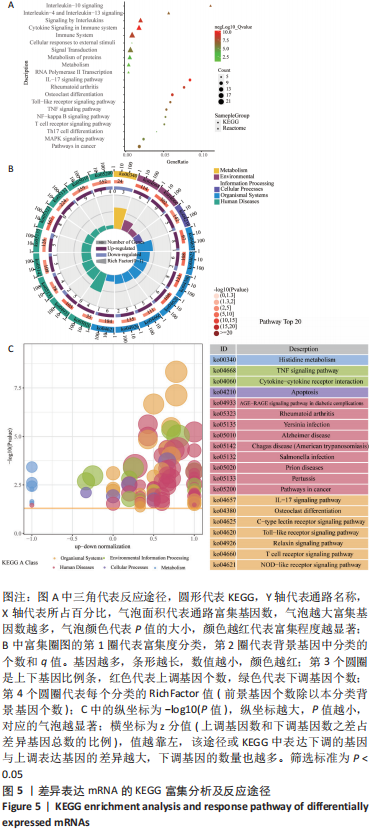
随后,运用KOBAS 3.0对差异表达mRNAs进行GO、KEGG和反应途径的富集分析,q < 0.05为条件,筛选出富集显著的GO条目和功能途径。结果显示,差异表达mRNA的GO富集分析中,生物过程(Biological process,BP)主要涉及炎症反应与调控、血管生成、细胞因子介导的信号通路、凋亡过程的负调控、细胞群增殖的正调控等,根据q < 0.05筛选出前10个GO条目绘制弦图和圈图可视化,见图4。KEGG通路富集分析显示,差异表达mRNA主要与细胞因子-细胞因子受体相互作用、破骨细胞分化、类风湿性关节炎以及IL-17信号通路、MAPK信号通路、Toll样受体信号通路、肿瘤坏死因子信号通路、NOD样受体信号通路、NF-κB信号通路、T细胞受体信号通路等免疫与炎症相关信号通路有关。反应途径表明差异表达mRNA主要富集在免疫系统中的细胞因子信号转导以及白细胞介素信号转导等途径。 "
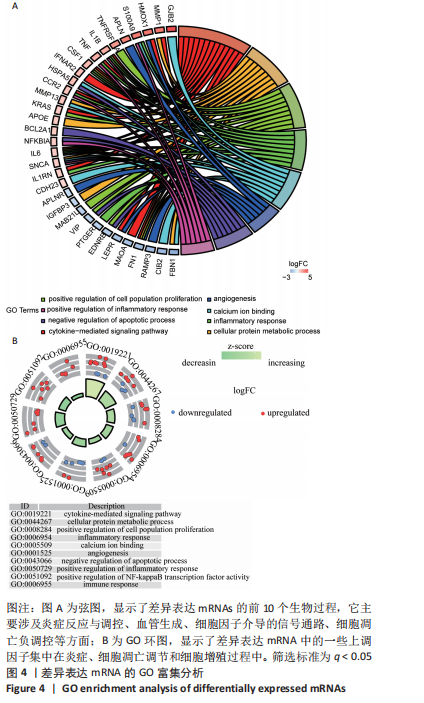

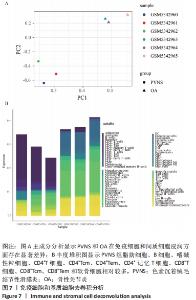
Spearman相关分析表明,见图8,IL-6、TNF与Macrophages M2(r=0.941,P=0.005)、cDC(r=0.928,P=0.008)、NKT(r=0.880,P=0.021)等呈正相关,与Tregs(r=-0.841,P=0.036)、GMP(r=-0.880,P=0.021)等呈负相关。CD163与CD4+Tem(r=0.986,P=0)、祖B细胞(r=0.941,P=0.005)、Th1细胞(r=0.845,P=0.034)、B细胞(r=0.845,P=0.034)呈正相关,与Plasma细胞(r=-0.820,P=0.046)、Adipocytes(r=-0.880,P=0.021)等呈负相关。CCR2与巨噬细胞M2(r=0.941,P=0.005)、自然杀伤T细胞(r=0.880,P=0.021)等呈正相关,与脂细胞、神经元等呈负相关(r=-0.880,P=0.021)。FN1与Macrophages M2(r=0.941,P=0.005)、cDC(r=0.928,P=0.008)等呈正相关,与Osteoblast(r=-0.880,P=0.021)、GMP(r=-0.880,P=0.021)等呈负相关。GAPDH与Macrophages M2(r=0.893,P=0.016)、CD4+Tem(r=0.897,P=0.015)、pro B细胞(r=0.893,P=0.016)成正相关,与Fibroblasts(r=-0.832,P=0.04)、Megakaryocytes(r=-0.832,P=0.04)等呈负相关。HMOX1与CD4+Tem(r=0.928,P=0.008)、Macrophages M2(r=0.880,P=0.021)等呈正相关,与Plasma细胞(r=-0.820,P=0.046) 、Fibroblasts(r=-0.986,P=0)等呈负相关。IL1B与Macrophages M2(r=0.880,P=0.021)、NKT(r=0.880,P=0.021)等呈正相关,与MEP(r=-0.833,P=0.039)、Megakaryocytes(r=-0.894,P=0.016)等呈负相关。KRAS与NKT(r=0.941,P=0.005)、aDC(r=0.845,P=0.034)等呈正相关,与Endothelial细胞(r=-0.941,P=0.005)、Chondrocytes(r=-0.845,P=0.034)等呈负相关。MAPK1与CD4+Tcm(r=0.845,P=0.034)、NKT(r=0.880,P=0.021)等呈正相关,与Preadipocytes(r=-0.829,P=0.042)、Megakaryocytes(r=-0.880,P=0.021)等呈负相关。NFKBI与Hepatocytes(r=0.941,P=0.005)、CD8+Tcm(r=0.841,P=0.036)等呈正相关,与Adipocytes、Neurons等呈负相关(r=-0.880,P=0.021)。PTPRC与NKT(r=0.941,P=0.005)、CD8+Tcm(r=0.928,P=0.008)等呈正相关,与MEP(r=-0.833,P=0.039)、Megakaryocytes(r=-0.880,P=0.021)等呈负相关。 "
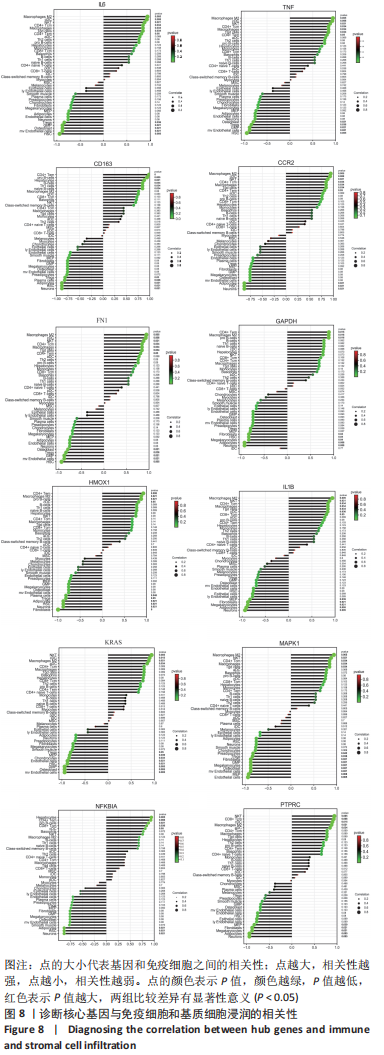

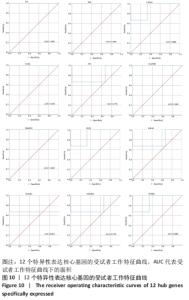
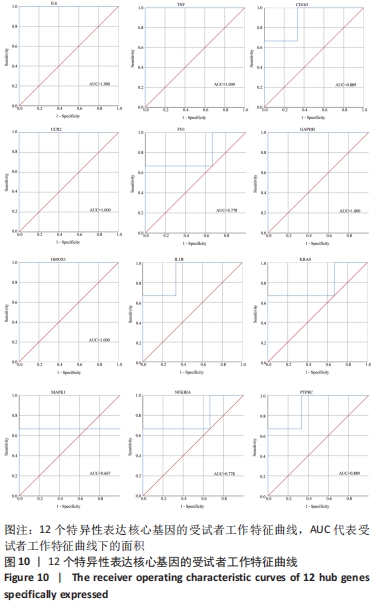
2.7 特异性表达核心基因的验证 选取GSE176133数据集(包含3个PVNS样本和3个骨性关节炎样本)来验证12个特异性表达核心基因。用Omicshare进行统计分析,采用t检验统计分析并绘制箱线图,见图9。统计结果与预测一致,PVNS样本中的12个特异性表达核心基因表达水平显著高于骨性关节炎样本中核心基因的表达水平(P < 0.05)。使用IBM SPSS (v26.0)对GSE176133中PVNS样本和骨性关节炎样本的12个特异性表达核心基因表达谱进行ROC分析,并绘制ROC曲线。ROC曲线下与坐标轴围成的面积被定义为Area Under Curve(AUC)。AUC越接近1.0,检测方法真实性越高;等于0.5时,则真实性最低,无应用价值[37]。通过分析结果发现,与骨性关节炎样本相比,这12个特异性表达核心基因在PVNS样本中具有较高的诊断价值。其中,TNF、IL-6、CCR2、GAPDH、HMOX1在PVNS样本中的诊断价值相对较高(AUC:1.000),其他核心基因对PVNS的诊断价值分别为:CD163(AUC:0.889)、FN1(AUC:0.778)、IL1B(AUC:0.889)、KRAS(AUC:0.778)、MAPK1(AUC:0.667)、NFKBIA(AUC:0.778)、PTPRC(AUC:0.889),见图10。由于它们在PVNS样本中表现出良好诊断性能,因此,根据目前的样本,推测这12个特异性表达核心基因可能是诊断PVNS的生物标志物。"
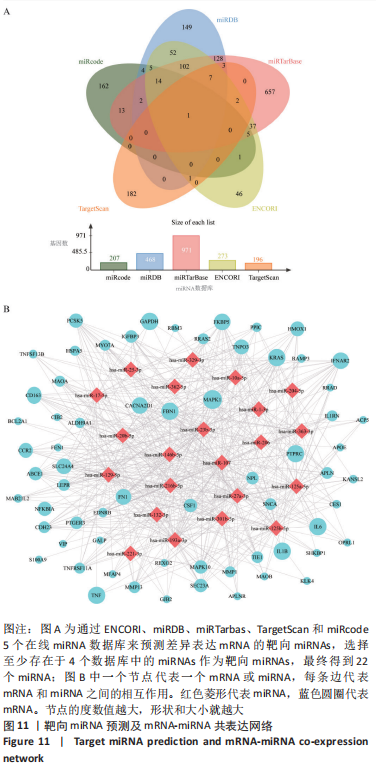
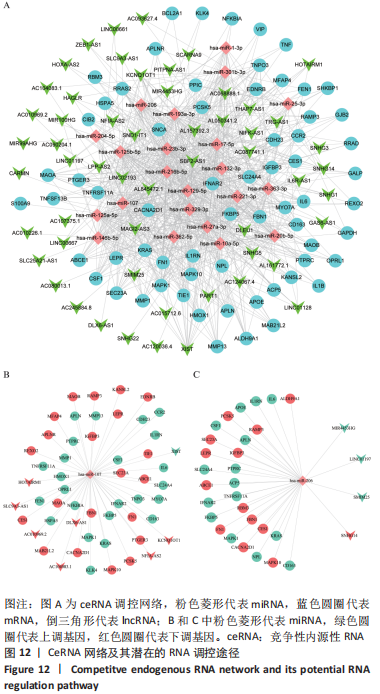
ceRNA 假说揭示了一种 RNA 间相互作用的新机制,miRNA作为一个转录后调控的重要因子,其活性可被 lncRNA通过“海绵”吸附的方式调控ceRNA。lncRNA作为ceRNA竞争性地与miRNA结合影响miRNA的功能,从而调节编码基因的蛋白质水平,参与靶基因的表达调控。使用ENCORI数据库预测与差异表达lncRNA相互作用的miRNA,筛选条件设定为:human,hg19基因组,high stringency≥3。最终获得3 676个miRNA-lncRNA对,运用Cytoscape软件构建lncRNA-miRNA表达网络。在 Cytoscape 软件中将miRNA-mRNA和IncRNA-miRNA表达网络合并,借助其“merge”功能得到IncRNA-miRNA-mRNA相互作用的ceRNA调控网络,见图12A。根据ceRNA假说,检索了大量文献发现,PVNS相关RNA研究较少,但PVNS相关ceRNA网络中包含的hsa-miRNA-206在骨性关节炎组织和软骨细胞中显著升高,并明显抑制软骨细胞增殖,促进细胞凋亡[38]。此外,hsa-miR-206已确定为多种肿瘤疾病中的重要抑制基因[39]。另一个包含在网络中的hsa-miR-107已被证明可抑制细胞增殖并调节脂质代谢和免疫炎症过程[40]。hsa-miR-107在骨性关节炎软骨细胞中的表达水平明显低于正常软骨细胞,miR-107的过表达可以抑制骨性关节炎软骨细胞的凋亡[41]。选取hsa-miR-206和hsa-miR-107建立子网络进一步分析,见图12B,C。根据mRNA、miRNA和lncRNA之间的上下调关系,作者认为SNHG14-hsa-miR-206-FN1和XIST-hsa-miR-107-HMOX1/CD163可能是调节PVNS疾病进展的潜在RNA调控途径。 "
| [1] TYLER WK, VIDAL AF, WILLIAMS RJ, et al. Pigmented villonodular synovitis. J Am Acad Orthop Surg. 2006;14(6):376-385. [2] GEIGER EV, REIZE P, RUDERT M, et al. Pigmented villonodular synovitis. MMW Fortschr Med. 2006;148(6):40-41. [3] COURT S, NISSEN MJ, GABAY C. Pigmented villonodular synovitis. Rev Med Suisse. 2014;10(421):609-610. [4] FAŁEK A, NIEMUNIS-SAWICKA J, WRONA K, et al. Pigmented villonodular synovitis. Folia Med Cracov. 2018;58(4):93-104. [5] XIE GP, JIANG N, LIANG CX, et al. Pigmented villonodular synovitis: a retrospective multicenter study of 237 cases. PLoS One. 2015;10(3): e0121451. [6] FANG Y, ZHANG Q. Recurrence of pigmented villonodular synovitis of the knee: A case report with review of literature on the risk factors causing recurrence. Medicine (Baltimore). 2020;99(16):e19856. [7] DÜRR HR, CAPELLEN CF, KLEIN A, et al. The effects of radiosynoviorthesis in pigmented villonodular synovitis of the knee. Arch Orthop Trauma Surg. 2019;139(5):623-627. [8] MOLLON B, LEE A, BUSSE JW, et al. The effect of surgical synovectomy and radiotherapy on the rate of recurrence of pigmented villonodular synovitis of the knee: an individual patient meta-analysis. Bone Joint J. 2015;97-B(4):550-557. [9] FECEK C, CARTER KR. Pigmented Villonodular Synovitis//StatPearls. Treasure Island (FL): StatPearls Publishing, 2021. [10] GOUIN F, NOAILLES T. Localized and diffuse forms of tenosynovial giant cell tumor (formerly giant cell tumor of the tendon sheath and pigmented villonodular synovitis). Orthop Traumatol Surg Res. 2017; 103(1S):S91-S97. [11] EFRIMA B, SAFRAN N, AMAR E, et al. Simultaneous pigmented villonodular synovitis and synovial chondromatosis of the hip: case report. J Hip Preserv Surg. 2018;5(4):443-447. [12] DYADYK OO, HRYHOROVSKA AV. Clinical and morphological correlations and histopathology of joint damage in patients with diffuse-type tenosynovial giant cell tumor. Wiad Lek. 2019;72(12 cz 1):2269-2276. [13] JAFFE HL, LICHTENSTEIN L, SUTRO CJ. Pigmented villonodular synovitis, bursitis and tenosynovitis: A discussion of the synovial and bursal equivalents of the tenosynovial lesion commonly denoted as xanthoma, xanthogranuloma, giant cell tumor or myeloplaxoma of tendon sheath with some consideration of this tendon sheath lesionitself. Arch Pathol 1941;31:731-765. [14] BERGER I, WECKAUF H, HELMCHEN B, et al. Rheumatoid arthritis and pigmented villonodular synovitis: comparative analysis of cell polyploidy, cell cycle phases and expression of macrophage and fibroblast markers in proliferating synovial cells. Histopathology. 2005; 46(5):490-497. [15] O’KEEFE RJ, ROSIER RN, TEOT LA, et al. Cytokine and matrix metalloproteinase expression in pigmented villonodular synovitis may mediate bone and cartilage destruction. Iowa Orthop J. 1998;18:26-34. [16] KROOT EJ, KRAAN MC, SMEETS TJ, et al. Tumour necrosis factor alpha blockade in treatment resistant pigmented villonodular synovitis. Ann Rheum Dis. 2005;64(3):497-499. [17] RITCHIE ME, PHIPSON B, WU D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [18] CAI W, LI H, ZHANG Y, et al. Identification of key biomarkers and immune infiltration in the synovial tissue of osteoarthritis by bioinformatics analysis. PeerJ. 2020;8:e8390. [19] TAO Z, SHI A, LI R, et al. Microarray bioinformatics in cancer- a review. J BUON. 2017;22(4):838-843. [20] SALMENA L, POLISENO L, TAY Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146(3):353-358. [21] BARRETT T, WILHITE SE, LEDOUX P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013; 41(Database issue):D991-D995. [22] ZHOU G, SOUFAN O, EWALD J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234-W241. [23] WANG Y, ZHANG S, LI F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031-D1041. [24] AMBERGER JS, BOCCHINI CA, SCHIETTECATTE F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789-D798. [25] STELZER G, ROSEN N, PLASCHKES I, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1-1.30.33. [26] WISHART DS, FEUNANG YD, GUO AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46: D1074-D1082. [27] PIÑERO J, BRAVO À, QUERALT-ROSINACH N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833-D839. [28] WU C, JIN X, TSUENG G, et al. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016; 44(D1):D313-D316. [29] SZKLARCZYK D, GABLE AL, LYON D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [30] BADER GD, HOGUE CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. [31] CHIN CH, CHEN SH, WU HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4(Suppl 4):S11. [32] SUBRAMANIAN A, KUEHN H, GOULD J, et al. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007; 23(23):3251-3253. [33] BU D, LUO H, HUO P, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49(W1):W317-W325. [34] ARAN D, HU Z, BUTTE AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. [35] JOLLIFFE IT, CADIMA J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374 (2065):20150202. [36] LI JH, LIU S, ZHOU H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92-D97. [37] CAO R, LÓPEZ-DE-ULLIBARRI I. ROC Curves for the Statistical Analysis of Microarray Data. Methods Mol Biol. 2019;1986:245-253. [38] NI Z, SHANG X, TANG G, et al. Expression of miR-206 in Human Knee Articular Chondrocytes and Effects of miR-206 on Proliferation and Apoptosis of Articular Chondrocytes. Am J Med Sci. 2018;355(3):240-246. [39] ZHANG H, WANG J, REN T, et al. Bone marrow mesenchymal stem cell-derived exosomal miR-206 inhibits osteosarcoma progression by targeting TRA2B. Cancer Lett. 2020;490:54-65. [40] ZHANG Z, WU S, MUHAMMAD S, et al. miR-103/107 promote ER stress-mediated apoptosis via targeting the Wnt3a/β-catenin/ATF6 pathway in preadipocytes. J Lipid Res. 2018;59(5):843-853. [41] ZHAO X, LI H, WANG L. MicroRNA-107 regulates autophagy and apoptosis of osteoarthritis chondrocytes by targeting TRAF3. Int Immunopharmacol. 2019;71:181-187. [42] SHEKHAR A, SINGH S, PATIL SS, et al. Osteochondral Lesion in Diffuse Pigmented Villonodular Synovitis of the Knee. Knee Surg Relat Res. 2019;31(1):67-71. [43] FECEK C, CARTER KR. Pigmented Villonodular Synovitis. In: StatPearls. Treasure Island (FL): StatPearls Publishing, 2022. [44] SUN R, TIAN Z, KULKARNI S, et al. IL-6 prevents T cell-mediated hepatitis via inhibition of NKT cells in CD4+ T cell- and STAT3-dependent manners. J Immunol. 2004;172(9):5648-5655. [45] NAGASHIMA H, ISHII N, SO T. Regulation of Interleukin-6 Receptor Signaling by TNF Receptor-Associated Factor 2 and 5 During Differentiation of Inflammatory CD4+ T Cells. Front Immunol. 2018;9: 1986. [46] POVOLERI GAM, LALNUNHLIMI S, STEEL KJA, et al. Anti-TNF treatment negatively regulates human CD4+ T-cell activation and maturation in vitro, but does not confer an anergic or suppressive phenotype. Eur J Immunol. 2020;50(3):445-458. [47] ZHOU WH, DU WD, LI YF, et al. The Overexpression of Fibronectin 1 Promotes Cancer Progression and Associated with M2 Macrophages Polarization in Head and Neck Squamous Cell Carcinoma Patients. Int J Gen Med. 2022;15:5027-5042. [48] NEES TA, ROSSHIRT N, REINER T, et al. Inflammation and osteoarthritis-related pain. Schmerz. 2019;33(1):4-12. [49] JAMAL J, ROEBUCK MM, LEE SY, et al. Modulation of the mechanical responses of synovial fibroblasts by osteoarthritis-associated inflammatory stressors. Int J Biochem Cell Biol. 2020;126:105800. [50] WALLACH D. The cybernetics of TNF: Old views and newer ones. Semin Cell Dev Biol. 2016;50:105-114. [51] GREISEN SR, MOLLER HJ, STENGAARD-PEDERSEN K, et al. Soluble macrophage-derived CD163 is a marker of disease activity and progression in early rheumatoid arthritis. Clin Exp Rheumatol. 2011; 29(4):689-692. [52] BERGER I, EHEMANN V, HELMCHEN B, et al. Comparative analysis of cell populations involved in the proliferative and inflammatory processes in diffuse and localised pigmented villonodular synovitis. Histol Histopathol. 2004;19(3):687-692. [53] OHASHI Y, UCHIDA K, FUKUSHIMA K, et al. Correlation between CD163 expression and resting pain in patients with hip osteoarthritis: Possible contribution of CD163+ monocytes/macrophages to pain pathogenesis. J Orthop Res. 2022;40(6):1365-1374. [54] WU HZ, XIAO JQ, XIAO SS, et al. KRAS: A Promising Therapeutic Target for Cancer Treatment. Curr Top Med Chem. 2019;19(23):2081-2097. [55] HAMARSHEH S, GROß O, BRUMMER T, et al. Immune modulatory effects of oncogenic KRAS in cancer. Nat Commun. 2020;11(1):5439. [56] WU Z, SHOU L, WANG J, et al. Identification of the key gene and pathways associated with osteoarthritis via single-cell RNA sequencing on synovial fibroblasts. Medicine (Baltimore). 2020;99(33):e21707. [57] YANG S, OHE R, AUNG NY, et al. Comparative study of HO-1 expressing synovial lining cells between RA and OA. Mod Rheumatol. 2021;31(1): 133-140. [58] SANADA Y, TAN SJO, ADACHI N, et al. Pharmacological Targeting of Heme Oxygenase-1 in Osteoarthritis. Antioxidants (Basel). 2021; 10(3):419. [59] ISHIHARA S, OBEIDAT AM, WOKOSIN DL, et al. The role of intra-articular neuronal CCR2 receptors in knee joint pain associated with experimental osteoarthritis in mice. Arthritis Res Ther. 2021;23(1):103. [60] WANG Z, WANG B, ZHANG J, et al. Chemokine (C-C Motif) Ligand 2/Chemokine Receptor 2 (CCR2) Axis Blockade to Delay Chondrocyte Hypertrophy as a Therapeutic Strategy for Osteoarthritis. Med Sci Monit. 2021;27:e930053. [61] DECLERCQ HA, FORSYTH RG, VERBRUGGEN A, et al. CD34 and SMA expression of superficial zone cells in the normal and pathological human meniscus. J Orthop Res. 2012;30(5):800-808. [62] KUBSIK-GIDLEWSKA A, KLUPIŃSKI K, KROCHMALSKI M, et al. CD34+ Stem Cell Treatment for Knee Osteoarthritis: A Treatment and Rehabilitation Algorithm. J Rehabil Med Clin Commun. 2018;3:1000012. [63] WANG B, LI J, TIAN F. Downregulation of lncRNA SNHG14 attenuates osteoarthritis by inhibiting FSTL-1 mediated NLRP3 and TLR4/NF-κB pathway through miR-124-3p. Life Sci. 2021;270:119143. [64] LIU Y, LIU K, TANG C, et al. Long non-coding RNA XIST contributes to osteoarthritis progression via miR-149-5p/DNMT3A axis. Biomed Pharmacother. 2020;128:110349. |
| [1] | Zhou Jin, Xu Zhen. Circulating microRNAs as potential biomarkers of exercise response and adaptation in the skeletal muscle [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(14): 2257-2265. |
| [2] | Tang Wenjing, Wu Siyuan, Yang Chen, Tao Xi. Inflammatory responses in post-stroke depression [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(8): 1278-1285. |
| [3] | Zhang Jing, Zhang Zhao. Effect of crown restoration and crown prosthesis on periodontal tissue and periodontal biomarkers [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(34): 5543-5552. |
| [4] | Wen Jianyun, Miao Lili, Guan Di, Liu Xuan, Chen Libai, Feng Xiaoqin, Xu Xiaoxiao, Liu Qiujun, Wu Xuedong, He Yuelin. Correlation of the level of plasma biomarkers with acute graft-versus-host disease in children [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(31): 5032-5039. |
| [5] | Liu Chao, Zeng Zhaomu, Wen Xichao, Wu Wensong, Sun Chengyuan, Zheng Kebin. Role and clinical application prospects of exosomal non-coding RNAs in the occurrence and development of glioma [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(24): 3928-3936. |
| [6] | Qian Xiaofen, Zeng Ping, Liu Jinfu, Chen Cai, Fan Siqi, Nong Jiao, Deng Wei. Comprehensive analysis of competing endogenous RNA network and potential biomarkers in alcohol-induced osteonecrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(23): 3670-3675. |
| [7] | Liu Xiaopeng, Zhang Sisen. Predictive value of blood biomarkers for brain injury after cardiac arrest [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(2): 302-307. |
| [8] | Yao Jiawei, Xu Xiongfeng, Yi Peng, Qiu Bo. Screening of differential genes and validation of key genes in synovial tissue of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(18): 2881-2887. |
| [9] | Liao Jianzhao, Xia Tian. The role of extracellular matrix in the occurrence and development of osteoarthritis and its clinical research value [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(12): 1937-1943. |
| [10] | Yuan Mei, Zhang Xinxin, Guo Yisha, Bi Xia. Diagnostic potential of circulating microRNA in vascular cognitive impairment [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1299-1304. |
| [11] | Lu Yuyun, Huang Mei, Shi Xinlei, Chen Baoyan. Bibliometric and visualization analysis of breast cancer stem cell literature from 2011 to 2020 based on Web of Science database [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(25): 4001-4008. |
| [12] | Lin Haiqi, Chen Liang, Tang Lu, Weng Xiquan, Lin Wentao. Significance of urinary proteomics assessing pathological changes in the body [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(20): 3259-3266. |
| [13] | Zhang Shuang, Tan Rui, Wang Chunxiao, Wu Fengyu, Guo Hongyu. MicroRNAs for assessing the motion control of human skeletal muscles [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(17): 2755-2760. |
| [14] |
Zeng Zhaomu, Wen Xichao, Zhang Yuhao, Geng Lianting, Shen Yang, Zheng Kebin.
Role and application of exosomes-mediated miRNAs in the treatment of glioma [J]. Chinese Journal of Tissue Engineering Research, 2020, 24(25): 4073-4080. |
| [15] | Yang Tianqi1, Hu Xiaowen1, Zhou Kejun2, Wan Chunling1. Plasma alpha-fetoprotein and other biochemical indicators for constructing early diagnosis models of potential biliary atresia in children [J]. Chinese Journal of Tissue Engineering Research, 2019, 23(31): 5046-5051. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||