[1] SOLIS EC, CARLIER IVE, KAMMINGA N, et al. Economic Evaluation of Self-Management for Patients with Persistent Depressive Disorder and their Caregivers. J Ment Health Policy Econ. 2024;27(4): 129-143.
[2] ZHANG ZQ, YANG MH, GUO ZP, et al. Increased prefrontal cortex connectivity associated with depression vulnerability and relapse. J Affect Disord. 2022;304:133-141.
[3] KESHAVARZ K, HEDAYATI A, REZAEI M, et al. Economic burden of major depressive disorder: a case study in Southern Iran. BMC Psychiatry. 2022; 22(1):577.
[4] ANDERSON E, CRAWFORD CM, FAVA M, et al. Depression - Understanding, Identifying, and Diagnosing. N Engl J Med. 2024;390(17):e41.
[5] ŁYSIK A, LOGOŃ K, SZCZYGIŁ A, et al. Innovative approaches in the treatment-resistant depression: exploring different therapeutic pathways. Geroscience. 2025. doi: 10.1007/s11357-025-01615-8.
[6] RAKEL RE. Depression. Prim Care. 1999; 26(2):211-224.
[7] FERKINGSTAD E, SULEM P, ATLASON BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021;53(12):1712-1721.
[8] FRESHOUR SL, KIWALA S, COTTO KC, et al. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144-D1151.
[9] QIU-QIANG Z, WEI-WEI Y, SHAN-SHU H, et al. Mendelian randomization of individual sleep traits associated with major depressive disorder. J Affect Disord. 2024;365:105-111.
[10] LI Y, LV MR, WEI YJ, et al. Dietary patterns and depression risk: A meta-analysis. Psychiatry Res. 2017;253:373-382.
[11] MENG X, NAVOLY G, GIANNAKOPOULOU O, et al. Multi-ancestry genome-wide association study of major depression aids locus discovery, fine mapping, gene prioritization and causal inference. Nat Genet. 2024;56(2):222-233.
[12] JAMET C, DUBERTRET C, LE STRAT Y, et al. Age of onset of major depressive episode and association with lifetime psychiatric disorders, health-related quality of life and impact of gender: A cross sectional and retrospective cohort study. J Affect Disord. 2024;363:300-309.
[13] BRUFFAERTS R, CAYWOOD K, AXINN WG. Early-life risk factors for depression among young adults in the United States general population: Attributable risks and gender differences. J Affect Disord. 2024;363:206-213.
[14] LIU S, XU D. Causal relationship between educational attainment and chronic pain: A Mendelian randomization study. Medicine (Baltimore). 2024;103(37): e39301.
[15] WANG Z, LI S, CAI G, et al. Mendelian randomization analysis identifies druggable genes and drugs repurposing for chronic obstructive pulmonary disease. Front Cell Infect Microbiol. 2024; 14:1386506.
[16] VÖSA U, CLARINGBOULD A, WESTRA HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021; 53(9):1300-1310.
[17] FINAN C, GAULTON A, KRUGER FA, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017; 9(383):eaag1166.
[18] SUN BB, CHIOU J, TRAYLOR M, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023; 622(7982):329-338.
[19] KELLY KM, SMITH JA, MEZUK B. Depression and interleukin-6 signaling: A Mendelian Randomization study. Brain Behav Immun. 2021;95:106-114.
[20] ZHOU H, JI Y, SUN L, et al. Exploring the causal relationships and mediating factors between depression, anxiety, panic, and atrial fibrillation: A multivariable Mendelian randomization study. J Affect Disord. 2024; 349:635-645.
[21] WANG Z, LIN X, CHEN X, et al. Genetic causality and metabolite pathway identifying the relationship of blood metabolites and psoriasis. Skin Res Technol. 2024;30(7):e13840.
[22] FAN M, YUN Z, YUAN J, et al. Genetic insights into therapeutic targets for gout: evidence from a multi-omics mendelian randomization study. Hereditas. 2024;161(1):56.
[23] STALEY JR, BLACKSHAW J, KAMAT MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016; 32(20):3207-3209.
[24] BURGESS S, DAVEY SMITH G, DAVIES NM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 2023;4:186.
[25] ZHANG C, HE Y, LIU L. Identifying therapeutic target genes for migraine by systematic druggable genome-wide Mendelian randomization. J Headache Pain. 2024;25(1):100.
[26] GIAMBARTOLOMEI C, VUKCEVIC D, SCHADT EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014; 10(5):e1004383.
[27] RASOOLY D, PELOSO GM, GIAMBARTOLOMEI C. Bayesian Genetic Colocalization Test of Two Traits Using coloc. Curr Protoc. 2022; 2(12):e627.
[28] GIAMBARTOLOMEI C, ZHENLI LIU J, ZHANG W, et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018; 34(15):2538-2545.
[29] IMPRIALOU M, PETRETTO E, BOTTOLO L. Expression QTLs Mapping and Analysis: A Bayesian Perspective. Methods Mol Biol. 2017;1488:189-215.
[30] HU S, LUO Y, ZHANG Z, et al. Protein function annotation based on heterogeneous biological networks. BMC Bioinformatics. 2022;23(1):493.
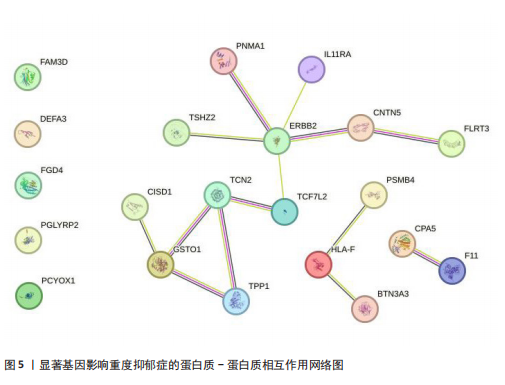
[31] SZKLARCZYK D, KIRSCH R, KOUTROULI M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023; 51(D1):D638-D646.
[32] KIM S, CHEN J, CHENG T, et al. PubChem 2023 update. Nucleic Acids Res. 2023; 51(D1):D1373-D1380.
[33] BERMAN HM, WESTBROOK J, FENG Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235-242.
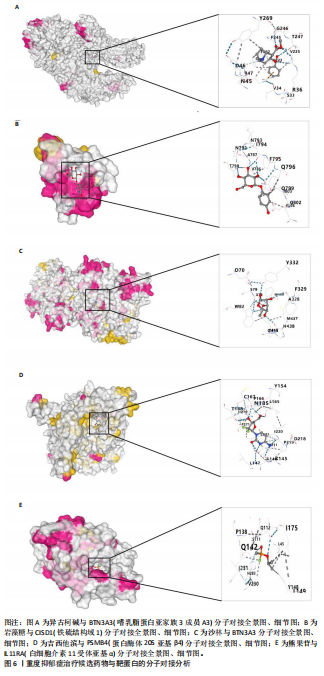
[34] MORRIS GM, HUEY R, LINDSTROM W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785-2791.
[35] WU H, HUANG J, ZHONG Y, et al. DrugSig: A resource for computational drug repositioning utilizing gene expression signatures. PLoS One. 2017; 12(5):e0177743.
[36] BHATTACHARYYA U, JOHN J, LAM M, et al. Circulating Blood-Based Proteins in Psychopathology and Cognition: A Mendelian Randomization Study. JAMA Psychiatry. 2025;82(5):481-491.
[37] TKACHEV A, STEKOLSHCHIKOVA E, VANYUSHKINA A, et al. Lipid Alteration Signature in the Blood Plasma of Individuals With Schizophrenia, Depression, and Bipolar Disorder. JAMA Psychiatry. 2023;80(3):250-259.
[38] MA C, ZHANG D, MA Q, et al. Arbutin inhibits inflammation and apoptosis by enhancing autophagy via SIRT1. Adv Clin Exp Med. 2021;30(5):535-544.
[39] WANG X, RIBEIRO C, NILSSON A, et al. Oxytocin Receptor Expression and Activation in Parasympathetic Brainstem Cardiac Vagal Neurons. eNeuro. 2025;12(8):ENEURO.0204-25.2025.
[40] CHEN B, CHEN Y, HE Z, et al. S146 and M148 within the mature chain domain of PSMB4 are crucial for degrading PRRSV nsp1α. Cell Mol Life Sci. 2025; 82(1):148.
[41] LV J, LIU G, WANG Z, et al. Internalized polystyrene nanoplastics trigger testicular damage and promote ferroptosis via CISD1 downregulation in mouse spermatocyte. J Nanobiotechnology. 2025;23(1):537.
[42] VERONA F, DI BELLA S, SCHIRANO R, et al. Cancer stem cells and tumor-associated macrophages as mates in tumor progression: mechanisms of crosstalk and advanced bioinformatic tools to dissect their phenotypes and interaction. Front Immunol. 2025;16:1529847.
[43] SI S, LIU H, XU L, et al. Identification of novel therapeutic targets for chronic kidney disease and kidney function by integrating multi-omics proteome with transcriptome. Genome Med. 2024;16(1):84.
[44] GERARDS WL, HOP FW, HENDRIKS IL, et al.Cloning and expression of a human pro(tea)some beta-subunit cDNA: a homologue of the yeast PRE4-subunit essential for peptidylglutamyl-peptide hydrolase activity. FEBS Lett. 1994;346(2-3):151-155.
[45] YANG C, YU P, YANG F, et al. PSMB4 inhibits cardiomyocyte apoptosis via activating NF-κB signaling pathway during myocardial ischemia/reperfusion injury. J Mol Histol. 2021;52(4):693-703.
[46] ZHANG X, LIN D, LIN Y, et al. Proteasome beta-4 subunit contributes to the development of melanoma and is regulated by miR-148b. Tumour Biol. 2017;39(6):1010428317705767.
[47] WANG H, HE Z, XIA L, et al. PSMB4 overexpression enhances the cell growth and viability of breast cancer cells leading to a poor prognosis. Oncol Rep. 2018;40(4):2343-2352.
[48] LI X, SHEN A, ZHAO Y, et al. Mendelian Randomization Using the Druggable Genome Reveals Genetically Supported Drug Targets for Psychiatric Disorders. Schizophr Bull. 2023;49(5):1305-1315.
[49] LIU J, CHENG Y, LI M, et al. Genome-wide Mendelian randomization identifies actionable novel drug targets for psychiatric disorders. Neuropsychopharmacology. 2023;48(2):270-280.
[50] HU J, FU J, CAI Y, et al. Bioinformatics and systems biology approach to identify the pathogenetic link of neurological pain and major depressive disorder. Exp Biol Med (Maywood). 2024;249:10129.
[51] DATTILO V, AMATO R, PERROTTI N, et al. The Emerging Role of SGK1 (Serum- and Glucocorticoid-Regulated Kinase 1) in Major Depressive Disorder: Hypothesis and Mechanisms. Front Genet. 2020; 11:826.
[52] JI D, LIU W, CUI X, et al. A receptor-like kinase SlFERL mediates immune responses of tomato to Botrytis cinerea by recognizing BcPG1 and fine-tuning MAPK signaling. New Phytol. 2023;240(3):1189-1201.
[53] BALHARA P, SHARMA S, VASUDEVA N, et al. Modulation of Nrf-2/HO-1/HIF-1α/TFAM pathways by Arbutin in rat model of cerebral ischemic stroke. Mol Cell Neurosci. 2025;134:104034.
[54] JAIN A, GOEL V, SONI S, et al. Comparative Efficacy and Toxicity of Modified FOLFIRINOX and Gemcitabine Plus Nab-Paclitaxel in Advanced Pancreatic Ductal Adenocarcinoma: A Real-World Retrospective Analysis. Cureus. 2025;17(7): e87389.
[55] ZHOU J, HU Y, JING P. Antidepressant-Like Effect of Saffron (Crocus sativus L.) in Mice Exposed to Chronic Unpredictable Mild Stress via Attenuating Neuroinflammation and Recovering Neuroplasticity. Mol Nutr Food Res. 2025:e70201. doi: 10.1002/mnfr.70201.
[56] SUDHOLZ H, MENG X, PARK HY, et al. Core fucosylation of IL-2RB is required for natural killer cell homeostasis. Cell Rep. 2025;44(8):116101.
[57] VOLNIN A, PARSIKOV A, TSYBULKO N, et al. Ergot alkaloid control in biotechnological processes and pharmaceuticals (a mini review). Front Toxicol. 2024;6:1463758.
|