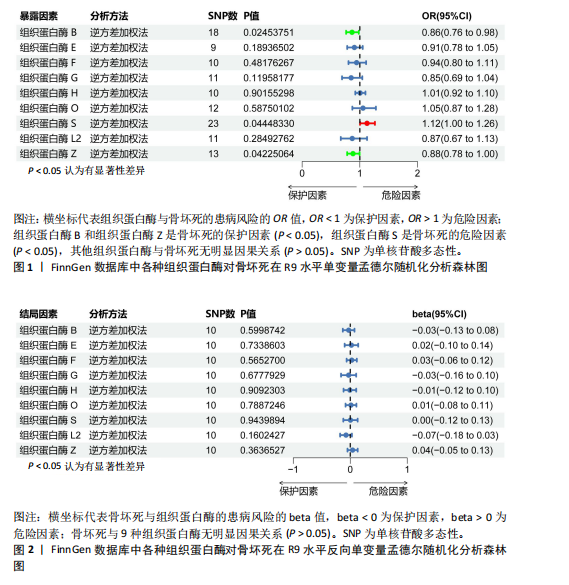
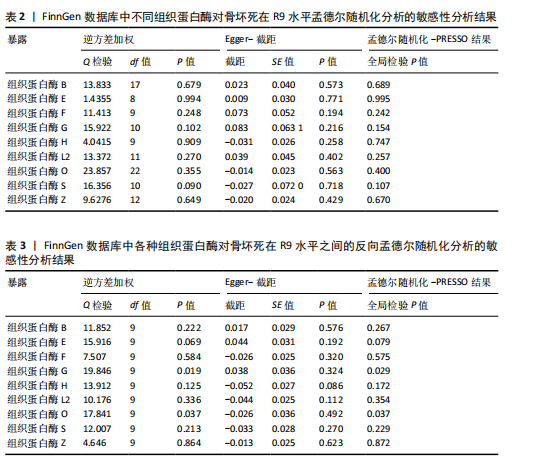
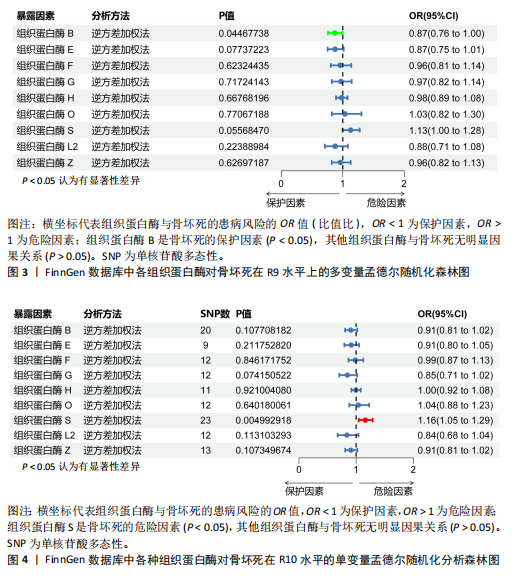
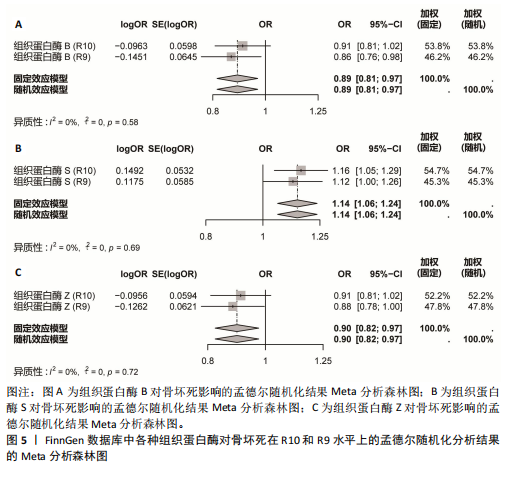
[1] MOTTA F, TIMILSINA S, GERSHWIN ME, et al. Steroid-induced osteonecrosis. J Transl Autoimmun. 2022;5:100168.
[2] BERGMAN J, NORDSTRÖM A, NORDSTRÖM P. Epidemiology of osteonecrosis among older adults in Sweden. Osteoporos Int. 2019;30(5):965-973.
[3] CHANG C, GREENSPAN A, GERSHWIN ME. The pathogenesis, diagnosis and clinical manifestations of steroid-induced osteonecrosis. J Autoimmun. 2020;110: 102460.
[4] REISER J, ADAIR B, REINHECKEL T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest. 2010;120(10): 3421-3431.
[5] PATEL S, HOMAEI A, EL-SEEDI HR, et al. Cathepsins: proteases that are vital for survival but can also be fatal. Biomed Pharmacother. 2018;105:526-532.
[6] RAUNER M, FÖGER-SAMWALD U, KURZ MF, et al. Cathepsin S controls adipocytic and osteoblastic differentiation, bone turnover, and bone microarchitecture. Bone. 2014; 64:281-287.
[7] ZHU G, CHEN W, TANG CY, et al. Knockout and Double Knockout of Cathepsin K and Mmp9 reveals a novel function of Cathepsin K as a regulator of osteoclast gene expression and bone homeostasis. Int J Biol Sci. 2022;18(14):5522-5538.
[8] CHOI JW, LIM S, JUNG SE, et al. Enhanced osteocyte differentiation: cathepsin d and l secretion by human adipose-derived mesenchymal stem cells. Cells. 2023; 12(24):2852.
[9] DUARTE C, YAMADA C, GARCIA C, et al. Crosstalk between dihydroceramides produced by Porphyromonas gingivalis and host lysosomal cathepsin B in the promotion of osteoclastogenesis. J Cell Mol Med. 2022;26(10):2841-2851.
[10] Birney E. Mendelian Randomization. Cold Spring Harb Perspect Med. 2022;12(4): a041302.
[11] SEKULA P, DEL GRECO MF, PATTARO C, et al. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016;27(11):3253-3265.
[12] LI W, CHAI JL, LI Z, et al. No evidence of genetic causality between diabetes and osteonecrosis: a bidirectional two-sample Mendelian randomization analysis. J Orthop Surg Res. 2023;18(1):970.
[13] HUANG X, DENG H, ZHANG B, et al. The causal relationship between cathepsins and digestive system tumors: a Mendelian randomization study. Front Oncol. 2024;14: 1365138.
[14] LIN L, WU Z, LUO H, et al. Cathepsin-mediated regulation of alpha-synuclein in Parkinson’s disease: a Mendelian randomization study. Front Aging Neurosci. 2024;16:1394807.
[15] YUSUFUJIANG A, ZENG S, LI H. Cathepsins and Parkinson’s disease: insights from Mendelian randomization analyses. Front Aging Neurosci. 2024;16:1380483.
[16] ZENG R, ZHOU Z, LIAO W, et al. Genetic insights into the role of cathepsins in cardiovascular diseases: a Mendelian randomization study. ESC Heart Fail. 2024; 11(5):2707-2718.
[17] CAO H, LIU B, GONG K, et al. Association between cathepsins and benign prostate diseases: a bidirectional two-sample Mendelian randomization study. Front Endocrinol (Lausanne). 2024;15:1348310.
[18] BOEF AG, DEKKERS OM, LE CESSIE S. Mendelian randomization studies: a review of the approaches used and the quality of reporting. Int J Epidemiol. 2015;44(2): 496-511.
[19] BURGESS S, LABRECQUE JA. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol. 2018; 33(10):947-952.
[20] SKRIVANKOVA VW, RICHMOND RC, WOOLF BAR, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR Statement. JAMA. 2021;326(16):1614-1621.
[21] SUN BB, MARANVILLE JC, PETERS JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73-79.
[22] KURKI MI, KARJALAINEN J, PALTA P, et al. A. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508-518.
[23] LI J, TANG M, GAO X, et al. Mendelian randomization analyses explore the relationship between cathepsins and lung cancer. Commun Biol. 2023;6(1):1019.
[24] GALA H, TOMLINSON I. The use of Mendelian randomisation to identify causal cancer risk factors: promise and limitations. J Pathol. 2020;250(5):541-554.
[25] KAMAT MA, BLACKSHAW JA, YOUNG R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851-4853.
[26] PIERCE BL, AHSAN H, VANDERWEELE TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740-752.
[27] VERBANCK M, CHEN CY, NEALE B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698.
[28] BURGESS S, BUTTERWORTH A, THOMPSON SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658-665.
[29] MOUNIER N, KUTALIK Z. Bias correction for inverse variance weighting Mendelian randomization. Genet Epidemiol. 2023; 47(4):314-331.
[30] BURGESS S, THOMPSON SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377-389.
[31] BOWDEN J, DAVEY SMITH G, HAYCOCK PC, et al. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016; 40(4):304-314.
[32] YAVORSKA OO, BURGESS S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734-1739.
[33] BOWDEN J, DEL GRECO MF, MINELLI C, et al. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36(11):1783-1802.
[34] HEMANI G, BOWDEN J, DAVEY SMITH G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195-R208.
[35] DELAISSÉ JM, EECKHOUT Y, VAES G. Inhibition of bone resorption in culture by inhibitors of thiol proteinases. Biochem J. 1980;192(1):365-368.
[36] UNEMORI EN, WERB Z. Collagenase expression and endogenous activation in rabbit synovial fibroblasts stimulated by the calcium ionophore A23187. J Biol Chem. 1988;263(31):16252-16259.
[37] AISA MC, RAHMAN S, SENIN U, et al. Cathepsin B activity in normal human osteoblast-like cells and human osteoblastic osteosarcoma cells (MG-63): regulation by interleukin-1 beta and parathyroid hormone. Biochim Biophys Acta. 1996;1290(1):29-36.
[38] CHAE HJ, HA KC, LEE GY, et al. Interleukin-6 and cyclic AMP stimulate release of cathepsin B in human osteoblasts. Immunopharmacol Immunotoxicol. 2007; 29(2):155-172.
[39] AISA MC, BECCARI T, COSTANZI E, et al. Biochim Biophys Acta. 2003;1621(2):149-159.
[40] PAGE AE, WARBURTON MJ, CHAMBERS TJ, et al. Human osteoclastomas contain multiple forms of cathepsin B. Biochim Biophys Acta. 1992;1116(1):57-66.
[41] RIFKIN BR, VERNILLO AT, KLECKNER AP, et al. Cathepsin B and L activities in isolated osteoclasts. Biochem Biophys Res Commun. 1991;179(1):63-69.
[42] RIFKIN BR, VERNILLO AT, KLECKNER AP, et al. Cathepsin B and L activities in isolated osteoclasts. Biochem Biophys Res Commun. 1991;179(1):63-69.
[43] BAUMGRASS R, WILLIAMSON MK, PRICE PA. Identification of peptide fragments generated by digestion of bovine and human osteocalcin with the lysosomal proteinases cathepsin B, D, L, H, and S. J Bone Miner Res. 1997;12(3):447-455.
[44] LAI WF, CHANG CH, TANG Y, et al. Early diagnosis of osteoarthritis using cathepsin B sensitive near-infrared fluorescent probes. Osteoarthritis Cartilage. 2004;12(3):239-244.
[45] BAICI A, HÖRLER D, LANG A, et al. Cathepsin B in osteoarthritis: zonal variation of enzyme activity in human femoral head cartilage. Ann Rheum Dis. 1995;54(4):281-288.
[46] BAICI A, LANG A, HÖRLER D, et al. Cathepsin B in osteoarthritis: cytochemical and histochemical analysis of human femoral head cartilage. Ann Rheum Dis. 1995;54(4):289-297.
[47] MEHRABAN F, TINDAL MH, PROFFITT MM, et al. Temporal pattern of cysteine endopeptidase (cathepsin B) expression in cartilage and synovium from rabbit knees with experimental osteoarthritis: gene expression in chondrocytes in response to interleukin-1 and matrix depletion. Ann Rheum Dis. 1997;56(2):108-115.
[48] ZHANG L, QIU J, SHI J, et al. MicroRNA-140-5p represses chondrocyte pyroptosis and relieves cartilage injury in osteoarthritis by inhibiting cathepsin B/Nod-like receptor protein 3. Bioengineered. 2021;12(2):9949-9964.
[49] MARTEL-PELLETIER J, CLOUTIER JM, PELLETIER JP. Cathepsin B and cysteine protease inhibitors in human osteoarthritis. J Orthop Res. 1990;8(3):336-344.
[50] BERARDI S, LANG A, KOSTOULAS G, et al. Alternative messenger RNA splicing and enzyme forms of cathepsin B in human osteoarthritic cartilage and cultured chondrocytes. Arthritis Rheum. 2001;44(8):1819-1831.
[51] HAMER I, DELAIVE E, DIEU M, et al. Up-regulation of cathepsin B expression and enhanced secretion in mitochondrial DNA-depleted osteosarcoma cells. Biol Cell. 2009;101(1):31-41.
[52] KRUEGER S, HAECKEL C, BUEHLING F, et al. Inhibitory effects of antisense cathepsin B cDNA transfection on invasion and motility in a human osteosarcoma cell line. Cancer Res. 1999;59(23):6010-6014.
[53] HAN SH, CHAE SW, CHOI JY, et al. Acidic pH environments increase the expression of cathepsin B in osteoblasts: the significance of ER stress in bone physiology. Immunopharmacol Immunotoxicol. 2009; 31(3):428-431. |