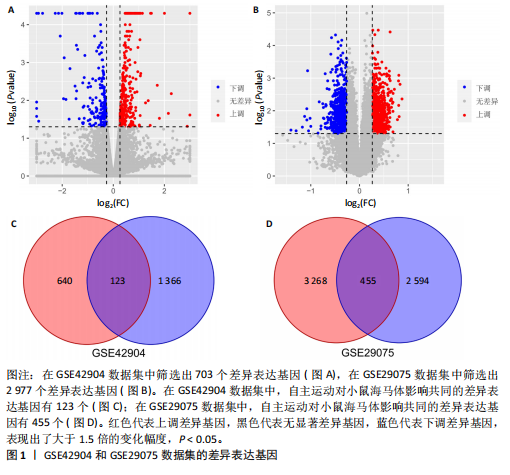
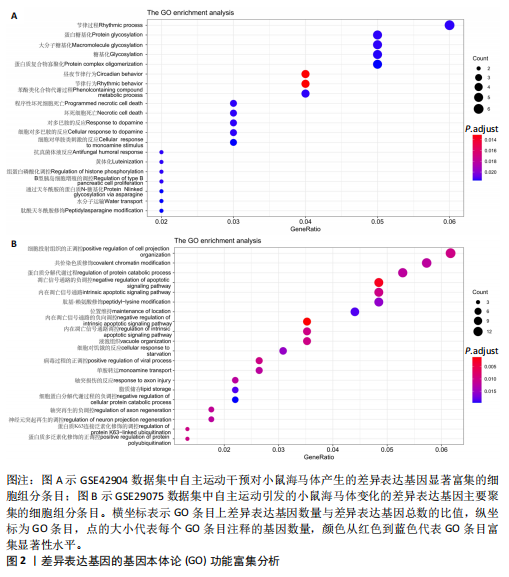
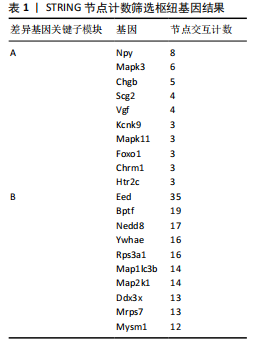
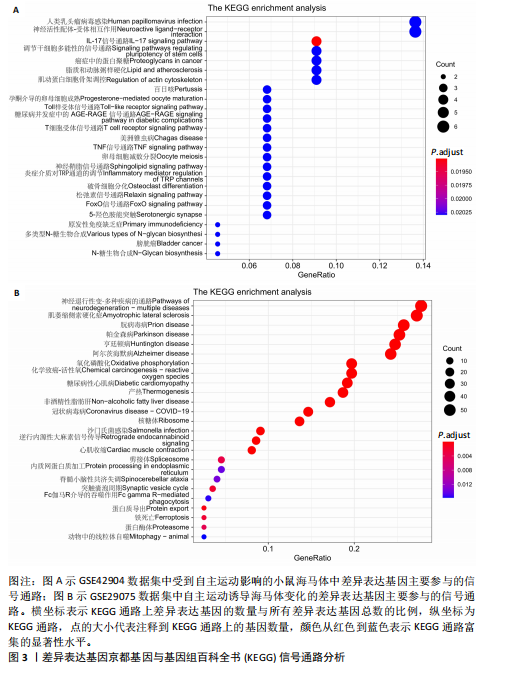
[1] PALAUS M, MARRON EM, VIEJO-SOBERA R, et al. Neural Basis of Video Gaming: A Systematic Review. Front Hum Neurosci. 2017;11:248.
[2] PLUVINAGE JV, WYSS-CORAY T. Systemic factors as mediators of brain homeostasis, ageing and neurodegeneration. Nat Rev Neurosci. 2020;21(2):93-102.
[3] VAN STRIEN NM, CAPPAERT NL, WITTER MP. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nat Rev Neurosci. 2009;10(4):272-282.
[4] WYSS-CORAY T, MUCKE L. Inflammation in neurodegenerative disease-a double-edged sword. Neuron. 2002;35(3):419-432.
[5] ZHANG YM, DAI QF, CHEN WH, et al. Effects of acupuncture on cortical expression of Wnt3a, β-catenin and Sox2 in a rat model of traumatic brain injury. Acupunct Med. 2016;34(1):48-54.
[6] BRAKSIEK M, THORMANN TF, WICKER P. Intentions of Environmentally Friendly Behavior Among Sports Club Members: An Empirical Test of the Theory of Planned Behavior Across Genders and Sports. Front Sports Act Living. 2021;3:657183.
[7] XU X, JERSKEY BA, COTE DM, et al. Cerebrovascular perfusion among older adults is moderated by strength training and gender. Neurosci Lett. 2014;560:26-30.
[8] BOLANDZADEH N, TAM R, HANDY TC, et al. Resistance Training and White Matter Lesion Progression in Older Women: Exploratory Analysis of a 12-Month Randomized Controlled Trial. J Am Geriatr Soc. 2015; 63(10):2052-2060.
[9] KENNEY WL, WILMORE JH, COSTILL DL. Physiology of sport and exercise. Human Kinetics, 2022.
[10] FORTE R, TOCCI N, DE VITO G. The Impact of Exercise Intervention with Rhythmic Auditory Stimulation to Improve Gait and Mobility in Parkinson Disease: An Umbrella Review. Brain Sci. 2021;11(6):685.
[11] DOS SANTOS JR, BORTOLANZA M, FERRARI GD, et al. One-Week High-Intensity Interval Training Increases Hippocampal Plasticity and Mitochondrial Content without Changes in Redox State. Antioxidants (Basel). 2020;9(5):445.
[12] CHO JW, JUNG SY, LEE SW, et al. Treadmill exercise ameliorates social isolation-induced depression through neuronal generation in rat pups. J Exerc Rehabil. 2017;13(6):627-633.
[13] JIANG H, CHEN S, WANG L, et al. An Investigation of Limbs Exercise as a Treatment in Improving the Psychomotor Speed in Older Adults with Mild Cognitive Impairment. Brain Sci. 2019;9(10):277.
[14] VOSS MW, NAGAMATSU LS, LIU-AMBROSE T, et al. Exercise, brain, and cognition across the life span. J Appl Physiol (1985). 2011;111(5):1505-1513.
[15] MA Q. Beneficial effects of moderate voluntary physical exercise and its biological mechanisms on brain health. Neurosci Bull. 2008;24(4):265-270.
[16] ANG ET, GOMEZ-PINILLA F. Potential therapeutic effects of exercise to the brain. Curr Med Chem. 2007;14(24):2564-2571.
[17] NAYLOR AS, BULL C, NILSSON MK, et al. Voluntary running rescues adult hippocampal neurogenesis after irradiation of the young mouse brain. Proc Natl Acad Sci U S A. 2008;105(38):14632-14637.
[18] NOTA MH, DOHM-HANSEN S, NICOLAS S, et al. A cafeteria diet blunts effects of exercise on adult hippocampal neurogenesis but not neurogenesis-dependent behaviours in adult male rats. BioRxiv. 2024: 2024.07. 16.603714.
[19] SEUDRE O, CARRILLO-BALTODANO AM, LIANG Y, et al. ERK1/2 is an ancestral organising signal in spiral cleavage. Nat Commun. 2022;13(1):2286.
[20] PATHAN M, KEERTHIKUMAR S, ANG CS, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015; 15(15):2597-2601.
[21] TERVO DG, HWANG BY, VISWANATHAN S, et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 2016;92(2):372-382.
[22] GAO Y, SYED M, ZHAO X. Mechanisms underlying the effect of voluntary running on adult hippocampal neurogenesis. Hippocampus. 2023;33(4):373-390.
[23] RAO YL, GANARAJA B, MURLIMANJU BV, et al. Hippocampus and its involvement in Alzheimer’s disease: a review. 3 Biotech. 2022;12(2):55.
[24] VIVAR C, PETERSON BD, VAN PRAAG H. Running rewires the neuronal network of adult-born dentate granule cells. Neuroimage. 2016;131:29-41.
[25] ZHU G, FANG Y, CUI X, et al. Magnolol upregulates CHRM1 to attenuate Amyloid-β-triggered neuronal injury through regulating the cAMP/PKA/CREB pathway. J Nat Med. 2022;76(1):188-199.
[26] LIU Z, GAO W, XU Y. Eleutheroside E alleviates cerebral ischemia-reperfusion injury in a 5-hydroxytryptamine receptor 2C (Htr2c)-dependent manner in rats. Bioengineered. 2022;13(5):11718-11731.
[27] YOU JC, MURALIDHARAN K, PARK JW, et al. Epigenetic suppression of hippocampal calbindin-D28k by ΔFosB drives seizure-related cognitive deficits. Nat Med. 2017; 23(11):1377-1383.
[28] HANON O, PEQUIGNOT R, SEUX ML, et al. Relationship between antihypertensive drug therapy and cognitive function in elderly hypertensive patients with memory complaints. J Hypertens. 2006;24(10):2101-2107.
[29] BARHWAL K, HOTA S K, BAITHARU I, et al. Isradipine antagonizes hypobaric hypoxia induced CA1 damage and memory impairment: Complementary roles of L-type calcium channel and NMDA receptors. Neurobiol Dis. 2009;34(2):230-244.
[30] CLAPHAM DE. Calcium signaling. Cell. 2007; 131(6):1047-58.
[31] MALIK BR, GILLESPIE JM, HODGE JJ. CASK and CaMKII function in the mushroom body α’/β’ neurons during Drosophila memory formation. Front Neural Circuits. 2013;7:52.
[32] NA KS, JUNG HY, KIM YK. The role of pro-inflammatory cytokines in the neuroinflammation and neurogenesis of schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2014;48:277-286.
[33] BEN-ARI S, OFEK K, BARBASH S, et al. Similar cation channels mediate protection from cerebellar exitotoxicity by exercise and inheritance. J Cell Mol Med. 2012;16(3):555-568.
[34] WAN C, SHI L, LAI Y, et al. Long-term voluntary running improves cognitive ability in developing mice by modulating the cholinergic system, antioxidant ability, and BDNF/PI3K/Akt/CREB pathway. Neurosci Lett. 2024;836:137872.
[35] CHISHOLM SP, CERVI AL, NAGPAL S, et al. Interleukin-17A increases neurite outgrowth from adult postganglionic sympathetic neurons. J Neurosci. 2012;32(4):1146-1155.
[36] PEARSON JRD, REGAD T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct Target Ther. 2017;2:17040.
[37] DRAGO A, SERRETTI A. Focus on HTR2C: A possible suggestion for genetic studies of complex disorders. Am J Med Genet B Neuropsychiatr Genet. 2009;150b(5):601-637.
[38] LIU F, LIANG C, LI Z, et al. Human iPSC-derived neuron of 16p11. 2 deletion reveals haplotype-specific expression of MAPK3 and its contribution to variable NDD phenotypes. BioRxiv. 2022: 2022.07.10.498576.
[39] GEIL CR, HAYES DM, MCCLAIN JA, et al. Alcohol and adult hippocampal neurogenesis: promiscuous drug, wanton effects. Prog Neuropsychopharmacol Biol Psychiatry. 2014;54:103-113.
[40] BIJAK M. Neuropeptide Y reduces epileptiform discharges and excitatory synaptic transmission in rat frontal cortex in vitro. Neurosci. 2000;96(3):487-494.
[41] DECRESSAC M, WRIGHT B, DAVID B, et al. Exogenous neuropeptide Y promotes in vivo hippocampal neurogenesis. Hippocampus. 2011;21(3):233-238.
[42] MALVA JO, XAPELLI S, BAPTISTA S, et al. Multifaces of neuropeptide Y in the brain--neuroprotection, neurogenesis and neuroinflammation. Neuropeptides. 2012; 46(6):299-308.
[43] STILLMAN CM, ESTEBAN-CORNEJO I, BROWN B, et al. Effects of Exercise on Brain and Cognition Across Age Groups and Health States. Trends Neurosci. 2020;43(7): 533-543.
[44] DI BENEDETTO G, IANNUCCI LF, SURDO NC, et al. Compartmentalized signaling in aging and neurodegeneration. Cells-Basel. 2021;10(2):464.
[45] DEN HEIJER T, VAN DER LIJN F, KOUDSTAAL PJ, et al. A 10-year follow-up of hippocampal volume on magnetic resonance imaging in early dementia and cognitive decline. Brain. 2010;133(Pt 4):1163-1172.
[46] MAASS A, DÜZEL S, GOERKE M, et al. Vascular hippocampal plasticity after aerobic exercise in older adults. Mol Psychiatry. 2015;20(5):585-593.
[47] ROSJAT N, LIU L, WANG BA, et al. Aging-associated changes of movement-related functional connectivity in the human brain. Neuropsychologia. 2018;117: 520-529.
[48] SPEISMAN RB, KUMAR A, RANI A, et al. Daily exercise improves memory, stimulates hippocampal neurogenesis and modulates immune and neuroimmune cytokines in aging rats. Brain Behav Immun. 2013;28: 25-43.
[49] VIVAR C, PETERSON B, PINTO A, et al. Running throughout Middle-Age Keeps Old Adult-Born Neurons Wired. eNeuro. 2023; 10(5):ENEURO.0084-23.2023.
[50] ZHANG Y, XU H. Identifying an APP-binding protein in neuronal cell death. Molecular Neurodegeneration. 2012;7(Suppl 1):L22.
[51] WANG YY, DENG YS, DAI SK, et al. Loss of microglial EED impairs synapse density, learning, and memory. Mol Psychiatry. 2022;27(7):2999-3009.
[52] BAHRAMPOUR S, JONSSON C, THOR S. Brain expansion promoted by polycomb-mediated anterior enhancement of a neural stem cell proliferation program. PLoS Biol. 2019;17(2):e3000163.
[53] KUEHNER JN, YAO B. The Dynamic Partnership of Polycomb and Trithorax in Brain Development and Diseases. Epigenomes. 2019;3(3):17-24. |