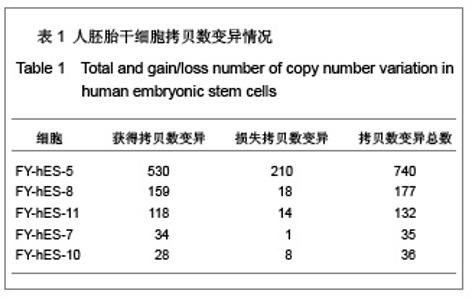
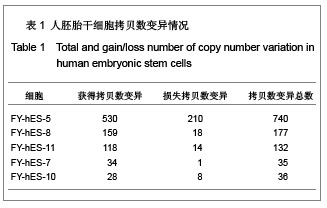
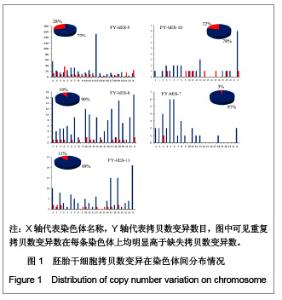
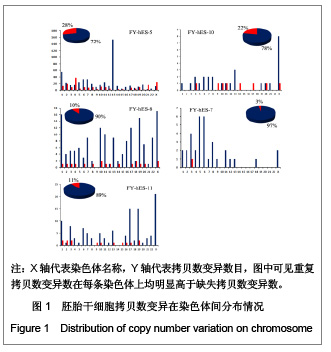
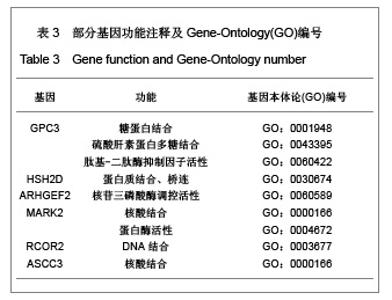
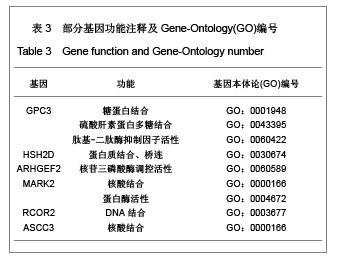
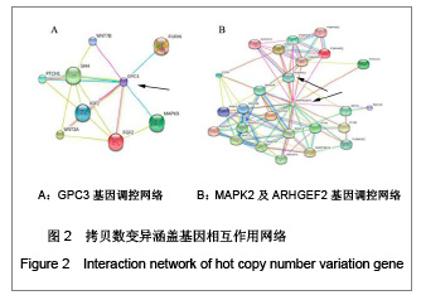
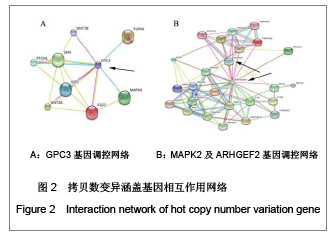
| [1] Baker DE, Harrison NJ, Maltby E,et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25(2):207-215.[2] Närvä E, Autio R, Rahkonen N,et al. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol. 2010;28(4):371-377.[3] Shen Y, Matsuno Y, Fouse SD,et al. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations.Proc Natl Acad Sci U S A. 2008;105(12):4709-4714.[4] Silva SS, Rowntree RK, Mekhoubad S,et al. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105(12):4820-4825.[5] Liu W, Sun X. Skewed X chromosome inactivation in diploid and triploid female human embryonic stem cells. Hum Reprod. 2009;24(8):1834-1843.[6] Sun X, Long X, Yin Y,et al. Similar biological characteristics of human embryonic stem cell lines with normal and abnormal karyotypes. Hum Reprod. 2008;23(10):2185-2193.[7] Yano S, Baskin B, Bagheri A,et al. Familial Simpson-Golabi-Behmel syndrome: studies of X-chromosome inactivation and clinical phenotypes in two female individuals with GPC3 mutations. Clin Genet. 2011; 80(5):466-471.[8] Minks J, Robinson WP, Brown CJ. A skewed view of X chromosome inactivation. J Clin Invest. 2008;118(1):20-23.[9] Kristiansen M, Langerød A, Knudsen GP,et al. High frequency of skewed X inactivation in young breast cancer patients. J Med Genet. 2002;39(1):30-33.[10] Ozbalkan Z, Bagi?lar S, Kiraz S,et al. Skewed X chromosome inactivation in blood cells of women with scleroderma. Arthritis Rheum. 2005;52(5):1564-1570.[11] Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS One. 2010;5(6):e11330.[12] Maitra A, Arking DE, Shivapurkar N,et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005; 37(10):1099-1103.[13] Hussein SM, Batada NN, Vuoristo S,et al. Copy number variation and selection during reprogramming to pluripotency.Nature. 2011;471(7336):58-62.[14] Diaz Perez SV, Kim R, Li Z,et al. Derivation of new human embryonic stem cell lines reveals rapid epigenetic progression in vitro that can be prevented by chemical modification of chromatin. Hum Mol Genet. 2012;21(4): 751-764.[15] Redon R, Ishikawa S, Fitch KR,et al. Global variation in copy number in the human genome. Nature. 2006;444(7118): 444-454.[16] Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61: 437-455.[17] Sebat J, Lakshmi B, Malhotra D,et al. Strong association of de novo copy number mutations with autism. Science. 2007; 316(5823):445-449.[18] Coe BP, Girirajan S, Eichler EE. The genetic variability and commonality of neurodevelopmental disease. Am J Med Genet C Semin Med Genet. 2012;160C(2):118-29.[19] Jobanputra V, Levy B, Kinney A,et al. Copy Number Changes on the X Chromosome in Women with and without Highly Skewed X-Chromosome Inactivation.Cytogenet Genome Res. 2012;136(4):264-269. |