Chinese Journal of Tissue Engineering Research ›› 2024, Vol. 28 ›› Issue (16): 2561-2567.doi: 10.12307/2024.321
Previous Articles Next Articles
CeRNA interaction network and immune manifestation of ferroptosis-related signature genes in rheumatoid arthritis
Xia Tian1, Li Binglin1, Xiao Fayuan1, Zheng Enze1, Chen Yueping2
- 1Guangxi University of Chinese Medicine, Nanning 530000, Guangxi Zhuang Autonomous Region, China; 2Department of Orthopedic Trauma and Hand Surgery, Ruikang Hospital, Guangxi University of Chinese Medicine, Nanning 530000, Guangxi Zhuang Autonomous Region, China
-
Received:2023-03-15Accepted:2023-05-10Online:2024-06-08Published:2023-07-31 -
Contact:Chen Yueping, MD, Chief physician, Doctoral supervisor, Department of Orthopedic Trauma and Hand Surgery, Ruikang Hospital, Guangxi University of Chinese Medicine, Nanning 530000, Guangxi Zhuang Autonomous Region, China -
About author:Xia Tian, MD candidate, Associate professor, Guangxi University of Chinese Medicine, Nanning 530000, Guangxi Zhuang Autonomous Region, China -
Supported by:National Natural Science Foundation of China, No. 81960803 (to CYP); Autonomous Region-level Doctoral Research Innovation Project of Guangxi University of Chinese Medicine, No. YCBXJ2021019 (to XT); Guangxi Traditional Chinese Medicine Appropriate Technology Development and Promotion Project, No. GZSY22-39 (to XT)
CLC Number:
Cite this article
Xia Tian, Li Binglin, Xiao Fayuan, Zheng Enze, Chen Yueping. CeRNA interaction network and immune manifestation of ferroptosis-related signature genes in rheumatoid arthritis[J]. Chinese Journal of Tissue Engineering Research, 2024, 28(16): 2561-2567.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

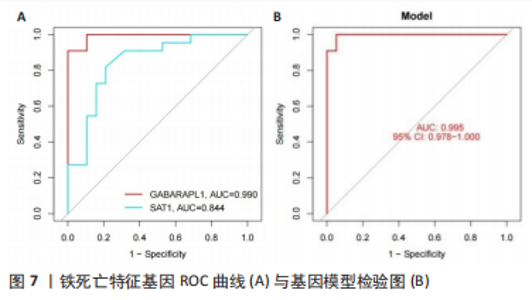
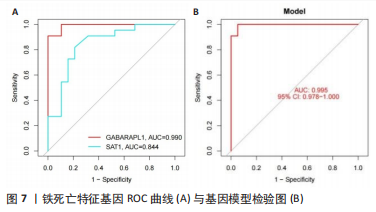
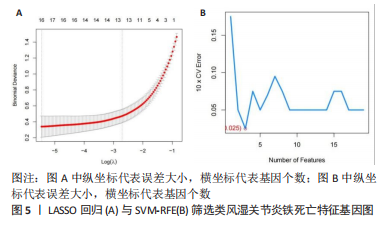
.3 机器学习方法筛选铁死亡疾病特征基因 运用LASSO回归方法,通过R软件中的“glmnet”对差异基因的表达量进行过滤,取误差最小值的前16 个基因为疾病特征基因,并绘制相关可视化图形,见图5A,基因误差按高到低排名依次获得SLC1A5、GABARAPL1、SAT1、PEBP1、MAPK8、MTCH1、SLC39A7、TRIM21、CIRBP、ADAM23、MEG3、RRM2、GDF15、AHCY、AR、BEX1;然后运用SVM-RFE方法筛选疾病特征基因,通过R软件中的“e1071、kernlab、caret”包对差异基因的表达量进行过滤,通过交叉验证寻找误差最小的疾病特征基因并做可视化分析,见图5B,发现误差表达最小的点共有3 个基因。接着将两者各自获得的疾病特征基因相交,并绘制VENN图,见图6,获得GABARAPL1、SAT1共2 个疾病特征基因。然后对获得的2 个疾病特征基因进行ROC曲线绘制,发现10 个疾病特征基因的AUC值都在80%以上,最后并对检测模型进行检测发现AUC值在0.95以上,说明GABARAPL1、SAT1为疾病的特征基因,具有一定的准确性,见图7。"

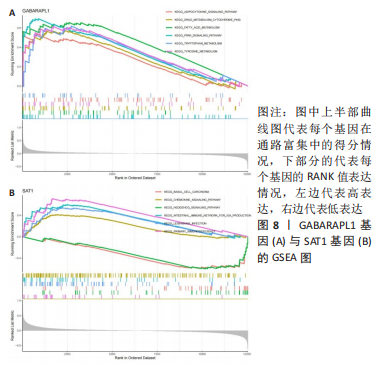
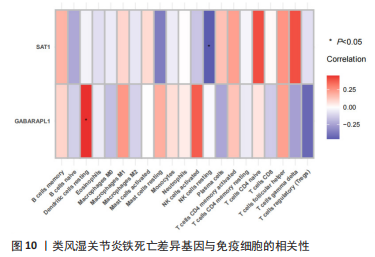
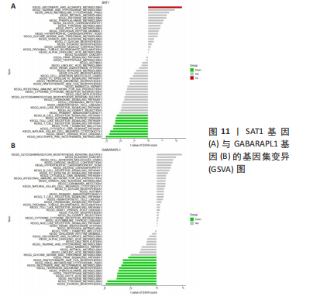
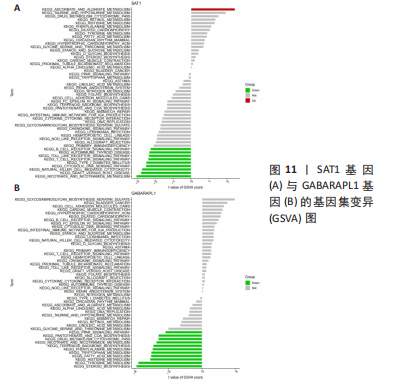
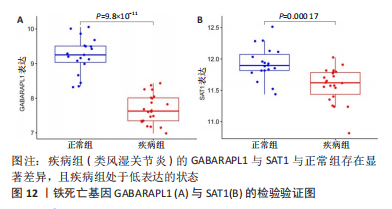
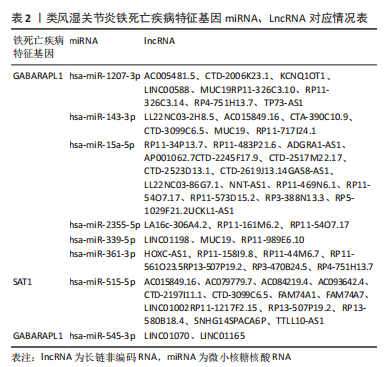
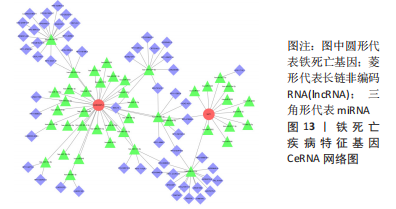
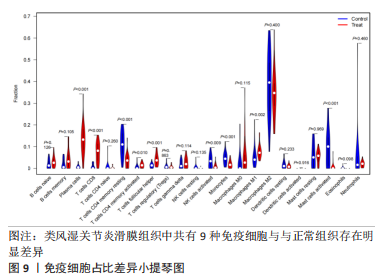
2.5 RA铁死亡疾病特征基因的免疫浸润分析 将矫正后的2 个芯片的基因表达量通过使用 CIBERSORT 算法进行分析,其中RA有22 个滑膜组织样本,正常组有19 个滑膜组织样本,同时筛选得到符合条件的有RA 22 个与正常组19 个之间的22 个免疫细胞的免疫浸润差异,通过与正常滑膜相比,RA滑膜中的浆细胞、T 细胞CD8、记忆静息T细胞CD4、记忆激活T细胞CD4、T细胞滤泡辅助器、激活的NK细胞、单核细胞、巨噬细胞M1、活化的肥大细胞等9种免疫细胞与正常组织存在明显差异,见图9,其中浆细胞、T细胞CD8、T细胞滤泡辅助器在疾病组中为高表达状态,其余为低表达。通过比较单个疾病特征基因与免疫细胞的相关性,发现GABARAPL1在树突状静息细胞、激活的NK细胞、巨噬细胞M1等为正相关,其中与树突状静息细胞的相关性最为显著,SAT1与T细胞CD4与γδT细胞为正相关,与NK静息细胞为负相关,见图10。通过GSVA分析发现SAT1在抗坏血酸和醛酸代谢上是上调的,而SAT1在B细胞受体信号通路、自身免疫性甲状腺疾病、TOLL样受体信号通路、T细胞受体信号通路、1型糖尿病、胞浆dna传感通路、自然杀伤细胞介导的细胞毒性、烟酸盐和烟酰胺的代谢是下调的;GABARAPL1在PPAR信号通路、泛酸与coa生物合成、药物代谢细胞色素P450、烟酸盐和烟酰胺的代谢、苯丙氨酸代谢、色氨酸代谢、脂肪酸代谢、类固醇生物合成等是下调的,见图11。通过将疾病特征基因对验证发现,疾病组的GABARAPL1与SAT1与正常组存在显著差异,且疾病组处于低表达的状态,这与以上的研究结果内容符合,见图12。"
| [1] OSIPOVA D, JANSSEN R, MARTENS HA. Rheumatoid arthritis: more than a joint disease. Ned Tijdschr Geneeskd. 2020;164:D4166. [2] MATEEN S, ZAFAR A, MOIN S, et al. Understanding the role of cytokines in the pathogenesis of rheumatoid arthritis. Clin Chim Acta. 2016;455:161-171. [3] GIANNINI D, ANTONUCCI M, PETRELLI F, et al. One year in review 2020: pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol. 2020;38(3):387-397. [4] XIE ZX, HOU HD, LUO D, et al. ROS-Dependent Lipid Peroxidation and Reliant Antioxidant Ferroptosis-Suppressor-Protein 1 in Rheumatoid Arthritis: a Covert Clue for Potential Therapy. Inflammation. 2021;44(1):35-47. [5] TANSKI W, CHABOWSKI M, JANKOWSKA-POLANSKA B, et al. Iron metabolism in patients with rheumatoid arthritis. Eur Rev Med Pharmacol Sci. 2021; 25(12):4325-4335. [6] SU LJ, ZHANG JH, GOMEZ H, et al. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxidative Med Cell Longev. 2019;2019:13. [7] DODSON M, CASTRO-PORTUGUEZ R, ZHANG DD. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019;23:7. [8] PHULL AR, NASIR B, UL HAQ I, et al. Oxidative stress, consequences and ROS mediated cellular signaling in rheumatoid arthritis. Chem-Biol Interact. 2018;281:121-136. [9] HUBER R, HUMMERT C, GAUSMANN U, et al. Identification of intra-group, inter-individual, and gene-specific variances in mRNA expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther. 2008;10(4):16. [10] WOETZEL D, HUBER R, KUPFER P, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther. 2014;16(2):21. [11] STOCKWELL BR, ANGELI JPF, BAYIR H, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171(2):273-285. [12] YANG GL, CHANG CC, YANG YW, et al. Resveratrol Alleviates Rheumatoid Arthritis via Reducing ROS and Inflammation, Inhibiting MAPK Signaling Pathways, and Suppressing Angiogenesis. J Agric Food Chem. 2018;66(49): 12953-12960. [13] FENG FB, QIU HY. Effects of Artesunate on chondrocyte proliferation, apoptosis and autophagy through the PI3K/AKT/mTOR signaling pathway in rat models with rheumatoid arthritis. Biomed Pharmacother. 2018;102:1209-1220. [14] WOJCIK P, GEGOTEK A, ZARKOVIC N, et al. Oxidative Stress and Lipid Mediators Modulate Immune Cell Functions in Autoimmune Diseases. Int J Mol Sci. 2021;22(2):20. [15] MAGTANONG L, KO PJ, TO M, et al. Exogenous Monounsaturated Fatty Acids Promote a Ferroptosis-Resistant Cell State. Cell Chem Biol. 2019;26(3):420-432.e9. [16] DIXON SJ, LEMBERG KM, LAMPRECHT MR, et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell. 2012;149(5):1060-1072. [17] YANG WS, STOCKWELL BR. Ferrootosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016;26(3):165-176. [18] GALARIS D, BARBOUTI A, PANTOPOULOS K. Iron homeostasis and oxidative stress: An intimate relationship. Biochim Biophys ActaMol Cell Res. 2019; 1866(12):15. [19] NAKAMURA T, NAGURO I, ICHIJO H. Iron homeostasis and iron-regulated ROS in cell death, senescence and human diseases. Biochim Biophys ActaGen Subj. 2019;1863(9):1398-1409. [20] CHOI RY, COYNER AS, KALPATHY-CRAMER J, et al. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl Vis Sci Technol. 2020;9(2):12. [21] LEE JM, LEE H, KANG S, et al. Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Biotechnological Advances. Nutrients. 2016;8(1):13. [22] MIRZA AZ, ALTHAGAFI II, SHAMSHAD H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur J Med Chem. 2019;166:502-513. [23] VENKATESH D, O’BRIEN NA, ZANDKARIMI F, et al. MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev. 2020;34(7-8):526-543. [24] MARION-LETELLIER R, SAVOYE G, GHOSH S. Fatty acids, eicosanoids and PPAR gamma. Eur J Pharmacol. 2016;785:44-49. [25] HU ZY, CHEN YD, ZHU SL, et al. Sonic Hedgehog Promotes Proliferation and Migration of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via Rho/ROCK Signaling. J Immunol Res. 2022;2022:9. [26] ZHU SL, YE YM, SHI YM, et al. Sonic Hedgehog Regulates Proliferation, Migration and Invasion of Synoviocytes in Rheumatoid Arthritis via JNK Signaling. Front Immunol. 2020;11:14. [27] LIU F, FENG XX, ZHU SL, et al. Sonic Hedgehog Signaling Pathway Mediates Proliferation and Migration of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via MAPK/ERK Signaling Pathway. Front Immunol. 2018;9:10. [28] WANG MX, ZHU SL, PENG WX, et al. Sonic Hedgehog Signaling Drives Proliferation of Synoviocytes in Rheumatoid Arthritis: A Possible Novel Therapeutic Target. J Immunol Res. 2014;2014:10. [29] HUANG SS, CHENG H, TANG CM, et al. Anti-oxidative, anti-apoptotic, and pro-angiogenic effects mediate functional improvement by sonic hedgehog against focal cerebral ischemia in rats. Exp Neurol. 2013;247:680-688. [30] ZHOU L, HUANG LT, XU QL, et al. Association of MUC19 Mutation With Clinical Benefits of Anti-PD-1 Inhibitors in Non-small Cell Lung Cancer. Front Oncol. 2021;11:12. [31] KERSCHNER JE, KHAMPANG P, ERBE CB, et al. Mucin gene 19 (MUC19) expression and response to inflammatory cytokines in middle ear epithelium. Glycoconjugate J. 2009;26(9):1275-1284. [32] TANG ZY, ZENG XH, LI JJ, et al. LncRNA HOXC-AS1 promotes nasopharyngeal carcinoma (NPC) progression by sponging miR-4651 and subsequently upregulating FOXO6. J Pharmacol Sci. 2021;147(3):284-293. [33] YU S, YU M, HE X, et al. KCNQ1OT1 promotes autophagy by regulating miR-200a/FOXO3/ATG7 pathway in cerebral ischemic stroke. Aging Cell. 2019;18(3):e12940. [34] ZHUANG SW, MA Y, ZENG YX, et al. METTL14 promotes doxorubicin-induced cardiomyocyte ferroptosis by regulating the KCNQ1OT1-miR-7-5p-TFRC axis. Cell Biol Toxicol. 2021. doi: 10.1007/s10565-021-09660-7. |
| [1] | Yu Weijie, Liu Aifeng, Chen Jixin, Guo Tianci, Jia Yizhen, Feng Huichuan, Yang Jialin. Advantages and application strategies of machine learning in diagnosis and treatment of lumbar disc herniation [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(9): 1426-1435. |
| [2] | Shen Jiangyong, He Xi, Tang Yuting, Wang Jianjun, Liu Jinyi, Chen Yuanyuan, Wang Xinyi, Liu Tong, Sun Haoyuan. RAS-selective lethal small molecule 3 inhibits the fibrosis of pathological scar fibroblasts [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(8): 1168-1173. |
| [3] | Bu Xianzhong, Bu Baoxian, Xu Wei, Zhang Chi, Zhang Yisheng, Zhong Yuanming, Li Zhifei, Tang Fubo, Mai Wei, Zhou Jinyan. Analysis of serum differential proteomics in patients with acute cervical spondylotic radiculopathy [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(4): 535-541. |
| [4] | Zhang Yaru, Chen Yanjun, Zhang Xiaodong, Chen Shenghua, Huang Wenhua. Effect of ferroptosis mediated by glutathione peroxidase 4 in the occurrence and progression of synovitis in knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(4): 550-555. |
| [5] | Ran Lei, Han Haihui, Xu Bo, Wang Jianye, Shen Jun, Xiao Lianbo, Shi Qi. Molecular docking analysis of the anti-inflammatory mechanism of Cibotium barometz and Epimedium for rheumatoid arthritis: animal experiment validation [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(2): 208-215. |
| [6] | Fan Yidong, Qin Gang, Su Guowei, Xiao Shifu, Liu Junliang, Li Weicai, Wu Guangtao. Establishment and analysis of osteoarthritis diagnosis model based on artificial neural networks [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(16): 2550-2554. |
| [7] | Han Weina, Xu Xiaoqing, Shi Jinning, Li Xinru, Cai Hongyan. Prediction and validation of potential targets for the glucagon-like peptide-1 receptor agonist in the treatment of Alzheimer’s disease [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(16): 2568-2573. |
| [8] | Xie Heng, Gu Ye, Gu Yingchu, Wu Zerui, Fang Tao, Wang Qiufei, Peng Yuqin, Geng Dechun, Xu Yaozeng. Ferroptosis in bone diseases: therapeutic targets of osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(16): 2613-2618. |
| [9] | Gao Zhengang, Zhang Xiaoyun, Jiang Wen. Mechanism of ferroptosis in osteoarthritis and its traditional Chinese medicine interventions [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(14): 2242-2247. |
| [10] | Yuan Tianyi, Liu Hongjiang, Yang Zengqiang, Lu Xingbao, Maimaitiyibubaji, Zhou Zhiheng, Cui Yong. Identification of differences in N6-methyladenosine-related genes in steroid-induced femoral head necrosis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(14): 2159-2165. |
| [11] | Zuo Jun, Ma Shaolin. Bioinformatics analysis and validation of differentially expressed genes and small molecule drug prediction in proliferative scar [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(14): 2166-2172. |
| [12] | Hu Mengfan, Yan Qiuhui, Deng Mengran, Liang Meimei, Liang Liang, Yi Sisi, Deng Jiagang, Yun Chenxia. Mangiferin inhibits proliferation, migration and inflammatory factor expression of fibroblast-like synoviocytes in rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(11): 1690-1695. |
| [13] | Yuan Changshen, Liao Shuning, Li Zhe, Guan Yanbing, Wu Siping, Hu Qi, Mei Qijie, Duan Kan. N6-methyladenosine related regulatory factors in osteoarthritis: bioinformatics analysis and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(11): 1724-1729. |
| [14] | Zeng Jiaxu, He Qi, Chen Bohao, Li Miao, Li Shaocong, Yang Junzheng, Pan Zhaofeng, Wang Haibin. An insight into the mechanism of iron overload in knee osteoarthritis under the theory of blood stasis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(11): 1743-1748. |
| [15] | Nong Fuxiang, Jiang Zhixiong, Li Yinghao, Xu Wencong, Shi Zhilan, Luo Hui, Zhang Qinglang, Zhong Shuang, Tang Meiwen. Bone cement augmented proximal femoral nail antirotation for type A3.3 intertrochanteric femoral fracturalysis [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(在线): 1-10. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||