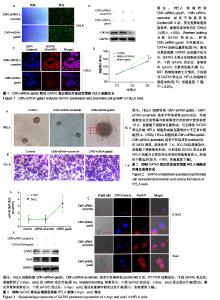
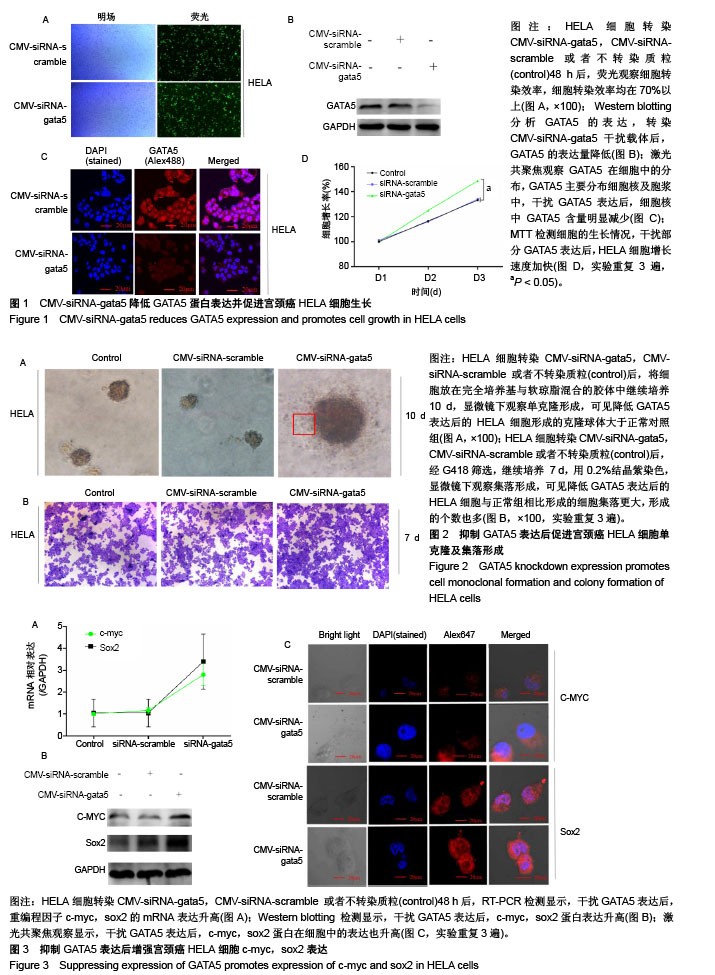
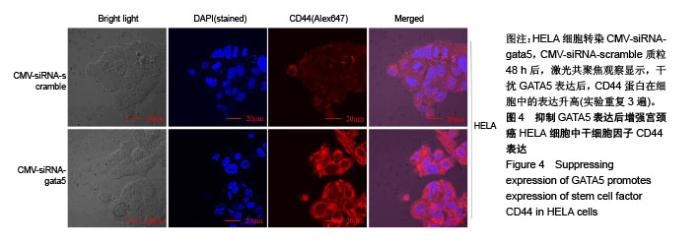
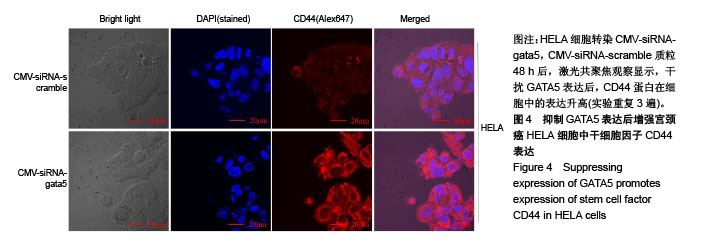
| [1] Li B, Yang XX, Wang D, et al. MicroRNA-138 inhibits proliferation of cervical cancer cells by targeting c-Met. Eur Rev Med Pharmacol Sci. 2016;20(6):1109-1114.[2] Feng D, Peng C, Li C, et al. Identification and characterization of cancer stem-like cells from primary carcinoma of the cervix uteri. Oncology Reports.2009;22(5):1129-1134.[3] Todaro M, Lombardo Y, Stassi G. Evidences of cervical cancer stem cells derived from established cell lines. Cell Cycle. 2010; 9(7):1231-1240.[4] Shu J, Zhang K, Zhang M, et al. GATA family members as inducers for cellular reprogramming to pluripotency. Cell Res. 2015;25(2):169-180.[5] Molkentin JD, Lin Q, Duncan SA, et al. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11(8):1061-1072.[6] Laverriere AC, Macneill C, Mueller C, et al. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem. 1994;269(37): 23177-23184. [7] Akiyama Y, Watkins N, Suzuki H, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer.Mol Cell Biol. 2003;23(23): 8429-8439.[8] Wen X, Akiyama Y, Zhou J, et al. Loss of transcription factor GATA-4/-5 by hypermethylation correlated with progression of gastric carcinomas. Cancer Res. 2007;67:2873-2873. [9] Lei X, Yan G, Zhang A, et al. Loss of GATA5 expression due to gene promoter methylation induces growth and colony formation of hepatocellular carcinoma cells. Oncol Lett. 2016; 11(1):861-869.[10] Liu P, Zhou TF, Qiu BA, et al. Methylation-Mediated Silencing of GATA5 Gene Suppresses Cholangiocarcinoma Cell Proliferation and Metastasis. Transl Oncol. 2018;11(3): 585-592.[11] Pei Y, Qin Y, Yuan S, et al. GATA4 promotes hepatoblastoma cell proliferation by altering expression of miR125b and DKK3. Oncotarget. 2016;7(47):77890-77901.[12] Lin B, Liu K, Wang W, et al.Expression and bioactivity of human α-fetoprotein in a Bac-to-Bac system.Biosci Rep. 2017;37(1). pii: BSR20160161.[13] Sun Y, Huang YH, Huang FY, et al. 3′-epi-12β-hydroxyfroside, a new cardenolide, induces cytoprotective autophagy via blocking the Hsp90/Akt/mTOR axis in lung cancer cells. Theranostics. 2018; 8(7):2044-2060. [14] Li M, Li H, Li C, et al. Cytoplasmic alpha-fetoprotein functions as a co-repressor in RA-RAR signaling to promote the growth of human hepatoma Bel 7402 cells. Cancer Lett. 2009;285(2): 190-199.[15] Li M, Zhu M, Li W, et al. Alpha-fetoprotein receptor as an early indicator of HBx-driven hepatocarcinogenesis and its applications in tracing cancer cell metastasis. Cancer Lett. 2013; 330(2):170-180.[16] Gandarillas A, Watt FM. c-Myc promotes differentiation of human epidermal stem cells. Genes Dev. 1997;11(21): 2869-2882.[17] Satoh Y, Matsumura I, Tanaka H, et al.Roles for c-Myc in self-renewal of hematopoietic stem cells.J Biol Chem. 2004; 279(24):24986-24993.[18] Martinez-Fernandez A, Nelson TJ, Ikeda Y, et al.c-MYC independent nuclear reprogramming favors cardiogenic potential of induced pluripotent stem cells.J Cardiovasc Transl Res. 2010;3(1):13-23.[19] Ellis P, Fagan BM, Magness ST, et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult.Dev Neurosci. 2004;26(2-4):148-165.[20] Giorgetti A, Montserrat N, Aasen T, et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5(4):353-357.[21] Park SB, Seo KW, So AY, et al. SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ. 2012; 19(3):534-545.[22] Xiang R, Liao D, Cheng T, et al. Downregulation of transcription factor SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer. Br J Cancer. 2011;104(9): 1410-1417.[23] Leis O, Eguiara A, Lopezarribillaga E, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31(11):1354-1365.[24] Bourguignon LY, Peyrollier K, Xia W, et al.Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells.J Biol Chem. 2008;283(25): 17635-17651.[25] Zhu H, Mitsuhashi N, Klein A, et al. The role of the hyaluronan receptor CD44 in mesenchymal stem cell migration in the extracellular matrix. Stem Cells.2010;24(4):928-935.[26] Yang X, Sarvestani SK, Moeinzadeh S, et al. Effect of CD44 binding peptide conjugated to an engineered inert matrix on maintenance of breast cancer stem cells and tumorsphere formation. Plos One. 2013;8(3):e59147.[27] Hong I, Hong SW, Chang YG, et al.Expression of the Cancer Stem Cell Markers CD44 and CD133 in Colorectal Cancer: An Immunohistochemical Staining Analysis.Ann Coloproctol. 2015; 31(3):84-91.[28] Moghbeli M, Moghbeli F, Forghanifard MM, et al. Cancer stem cell detection and isolation. Med Oncol. 2014;31(9):69.[29] Maddox J, Shakya A, South S, et al. Transcription factor Oct1 is a somatic and cancer stem cell determinant. Plos Genetics. 2012; 8(11):e1003048. |