Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (18): 4737-4748.doi: 10.12307/2026.751
Previous Articles Next Articles
Mechanism by which exercise regulates autophagy in different physiological systems
Zhang Shuli1, Hou Chaowen1, Yuan Shanshan2, Ma Yuhua3
- 1Qilu Insitute of Technology, Jinan 250200, Shandong Province, China; 2Binzhou Public Fitness Service Center, Binzhou 256600, Shandong Province, China; 3Sports and Leisure College, Shandong Sport University, Jinan 250102, Shandong Province, China
-
Received:2025-08-14Accepted:2025-09-19Online:2026-06-28Published:2025-12-08 -
Contact:Ma Yuhua, MD, Professor, Sports and Leisure College, Shandong Sport University, Jinan 250102, Shandong Province, China -
About author:Zhang Shuli, MS, Lecturer, Qilu Insitute of Technology, Jinan 250200, Shandong Province, China -
Supported by:Shandong Province Social Science Planning and Research Project, No. 17CTYJ04 (to MYH)
CLC Number:
Cite this article
Zhang Shuli, Hou Chaowen, Yuan Shanshan, Ma Yuhua . Mechanism by which exercise regulates autophagy in different physiological systems[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(18): 4737-4748.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

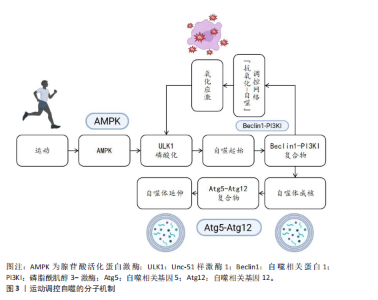
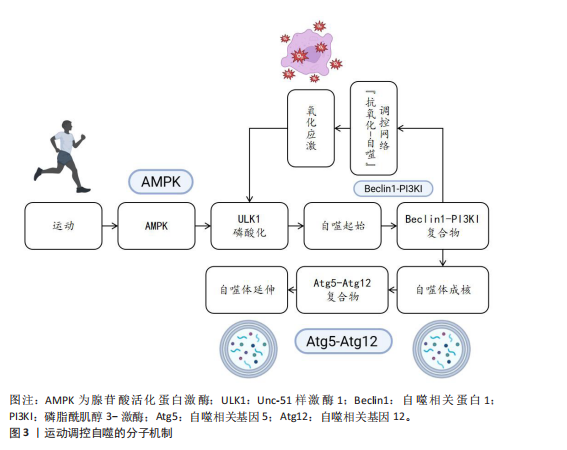
2.1 自噬概述 自噬是真核生物的一种高度保守的细胞内降解途径,选择性地降解细胞器、变性蛋白以及代谢废物,在细胞稳态及能量循环代谢中起着重要作用[1]。自噬可以分为巨自噬(吞噬膜吞噬胞质内容物形成自噬小体)、分子伴侣介导的自噬以及微自噬几种类型[7],其中巨自噬是调控运动的主要形式。自噬过程由Unc-51样激酶1复合体启动,经贝克林1-Ⅲ型磷脂酰肌醇-3-羟激酶复合体介导的吞噬膜成核、微管相关蛋白1轻链3Ⅱ依赖的自噬体延伸[2],再与溶酶体融合介导内容物降解,最后将释放出来的氨基酸、脂肪酸等可重新参与合成代谢[8]。运动通过能量应激(胰岛素样生长因子1/雷帕霉素靶蛋白信号)、氧化还原信号(核因子E2相关因子2/叉头框蛋白O)以及机械应激等多层次网络调节自噬活动[3],特别是腺苷酸活化蛋白激酶对自噬启动激酶1丝氨酸位点的磷酸化以及通过结节性硬化复合物2-Ras同源蛋白富集于脑蛋白调控轴对雷帕霉素靶蛋白复合物1的抑制是主要的分子开关[4],也可以通过运动的间歇性氧化应激反应激活PTEN诱导激酶1-帕金蛋白通路,进而增强线粒体自噬,从而起到控制细胞器质量、代谢重塑的双重作用[6]。表1介绍了运动调控自噬的关键时间点及重要内容。 2.2 运动调控自噬的分子机制 运动作为维持机体稳态的重要生理性刺激因素,通过多层次分子调控网络精准调控自噬过程。自噬作为真核生物中高度保守的细胞降解机制,可有效清除受损细胞器、错误折叠蛋白及代谢废物,它在细胞质量控制和能量代谢调节中发挥核心作用。运动调控自噬的分子机制见图3。 2.2.1 腺苷酸活化蛋白激酶通路 作为细胞能量稳态感知器,腺苷酸活化蛋白激酶在自噬激活中起到双重调控作用。当运动使腺苷三磷酸消耗引发腺苷一磷酸/腺苷三磷酸比值上升时,腺苷酸活化蛋白激酶经由肝激酶B1依赖性的磷酸化级联反应被活化,从两方面启动自噬过程。 一方面,腺苷酸活化蛋白激酶通过抑制雷帕霉素靶蛋白复合物1信号轴发挥调控作用。腺苷酸活化蛋白激酶直接磷酸化雷帕霉素靶蛋白复合物1的调控亚基相关蛋白(丝氨酸792位)和结节性硬化症蛋白2(苏氨酸1 227位/丝氨酸1 345位),进而抑制雷帕霉素靶蛋白复合物1催化亚基雷帕霉素靶蛋白的激酶活性[17,22]。这一过程解除了雷帕霉素靶蛋白复合物1对自噬启动激酶1复合物的抑制性磷酸化,促使自噬启动激酶1与自噬相关基因13、黏着斑激酶家族相互作用蛋白-200 kDa 组装形成具有功能活性的自噬起始复合体[23];另一方面,腺苷酸活化蛋白激酶可通过直接激活Unc-51样激酶1复合物发挥调控作用。腺苷酸活化蛋白激酶通过磷酸化Unc-51样激酶1的丝氨酸555、苏氨酸574 等核心位点,增强其激酶活性并驱动自噬小体成核[24]。此外,腺苷酸活化蛋白激酶还可通过上调自噬相关基因1启动子区域组蛋白H3第4位赖氨酸三甲基化修饰水平,促进自噬相关基因1、微管相关蛋白1轻链3B等自噬相关基因的转录表达,在翻译水平形成正向调控环路[25]。 尽管腺苷酸活化蛋白激酶介导的能量应激信号通路是自噬启动的核心通路,但它们在不同运动模式下的时空激活动力学的差异以及腺苷一磷酸活化蛋白激酶α1/α亚基2亚型在各组织自噬调控中的功能异质性,仍需通过单细胞磷酸化蛋白质组学技术进一步阐明。 2.2.2 雷帕霉素靶蛋白通路 雷帕霉素靶蛋白作为整合营养、生长因子、"

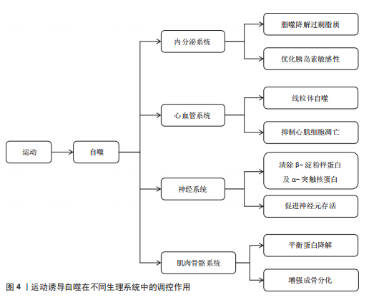
能量信号的中心激酶,它的活性抑制是运动诱导自噬的关键步骤。静息状态下,雷帕霉素靶蛋白复合物1通过磷酸化Unc-51样激酶1丝氨酸757、自噬相关基因13丝氨酸259等位点保持自噬抑制状态;而运动相关能量耗竭(腺苷酸活化蛋白激酶的活化)以及低氧信号可下调雷帕霉素靶蛋白复合物1的活性[12,26]。运动诱导的雷帕霉素靶蛋白抑制方式呈现出双相性,短期急性运动主要通过腺苷酸活化蛋白激酶-结节性硬化症蛋白2途径迅速抑制雷帕霉素靶蛋白,而长期适应性的运动依赖于生长因子信号的不断下调[27]。然而,雷帕霉素靶蛋白复合物1下游底物在运动诱导自噬中的特异性作用以及雷帕霉素靶蛋白复合物2在自噬体成熟阶段的调控尚有待挖掘。 2.2.3 自噬核心蛋白 在核心蛋白协同表达及翻译后修饰过程中,高效的自噬过程主要取决于核心蛋白及翻译后修饰,自噬相关基因1是核心蛋白之一,其复合物组装和激活在自噬过程中起重要作用[28]。自噬相关基因1是自噬起始复合物液泡分选蛋白34-液泡分选蛋白15-p150蛋白的重要组成部分,它的表达在运动后明显上升。研究表明,运动诱导过氧化物酶体增殖物激活受体γ辅激活因子1α通过结合自噬相关基因1基因启动子区域的雌激素相关受体α反应元件,上调自噬相关基因1在转录水平的合 成[28];自噬相关基因1的赖氨酸307位乙酰化修饰也能够阻止自噬相关基因1与自噬相关基因14的结合,而运动通过激活脱乙酰酶沉默信息调节因子 1逆转这种相互作用,促进自噬体成核[11]。此外,自噬依赖于微管相关蛋白轻链3的脂化与自噬体膜延伸过程。微管相关蛋白微管相关蛋白轻链3从胞质型微管相关蛋白1轻链3-Ⅰ到膜结合型微管相关蛋白1轻链3-Ⅱ的转化,是自噬体形成的标志。运动通过腺苷酸活化蛋白激酶依赖性磷酸化自噬相关蛋白7和自噬相关蛋白3,显著提升微管相关蛋白1轻链3-Ⅱ的脂化效率[29]。微管相关蛋白1轻链3-Ⅱ不仅是自噬活性的指标,也通过结合自噬相关基因8家族互作基序介导选择性清除受损线粒体,促进线粒体自噬,这一过程在运动诱导的心肌保护中尤为重要[11]。同样,增强自噬相关基因5-自噬相关基因12复合体功能对于自噬过程中也十分重要[29]。自噬相关基因5与自噬相关基因12的共价结合体在自噬延伸期作为核心成分,它的表达于运动后24 h达高峰,并与自噬体-溶酶体融合效率呈正相关[29]。虽然运动对于自噬关键蛋白表达调控已被明确,但其动态修饰对于自噬体成熟及底物选择的时空特性仍需依靠超高分辨率显微镜及活体动态追踪来探究。 2.2.4 氧化应激和自噬的关联 运动诱导的氧化应激是激活自噬的重要环境刺激因素,活性氧在其中也扮演着双重角色。高强度运动时,线粒体呼吸链电子泄露导致活性氧(超氧阴离子、过氧化氢)急剧升高,过多的活性氧可以直接损害DNA和蛋白质(如羰基化修饰),但是低水平的活性氧具有第二信使功能,激活p38丝裂原活化蛋白激酶/Unc-51样激酶1信号途径诱导选择性的自噬,以清除受损线粒体(线粒体自噬)[30]。在此过程中,核因子E2相关因子2通路发挥了重要功能:运动诱导核因子E2相关因子2进入细胞核,联合调控锰超氧化物歧化酶、过氧化氢酶等抗氧化基因和自噬相关基因16样蛋白1、自噬相关基因1等自噬基因的表达,形成“氧化应激-抗氧化-自噬”三角调控环路[31]。另外,抗氧化酶和自噬存在协同作用,超氧化物岐化酶家族通过减少活性氧发挥细胞保护功能,而锰超氧化物歧化酶缺失将明显抑制运动诱导的自噬激活,表明线粒体氧化损伤信号是启动自噬的必要条件[32]。此外,运动诱导过氧化氢酶减少过氧化氢水平,从而避免过氧化氢酶导致自噬相关基因蛋白的氧化失活,间接维持自噬通量,但活性氧的暴露强度和持续时间如何从启动自噬的信号转变为自噬抑制性损伤尚不明确。核因子E2相关因子2/胰岛素样生长因子1/雷帕霉素靶蛋白通路之间的时空维度交叉对话精准调控机制,仍然是运动生理学与细胞自噬学界的研究热点及未知科学问题。 2.3 运动诱导自噬在不同生理系统中的调控作用 不同生理系统对自噬的调控作用呈现明显差异,这些差异既与自噬的分子机制紧密关联,也受各系统特有生理需求和环境信号的影响。运动诱导自噬在不同生理系统中的调控作用见图4。 2.3.1 运动诱导自噬在内分泌系统中的调控作用 运动诱导的自噬过程,特别是脂质自噬、线粒体自噬,在调节内分泌的稳态方面具有重要作用。自噬过程通过选择性去除多余脂质和受损的线粒体降低氧化应激水平、改善线粒体功能,从而增加糖脂代谢水平[33],运动通过促进脂肪酸的氧化及降低机体中脂肪的堆积增强胰岛素敏感性,维持机体整体代谢平衡,从而维持机体内分泌稳态[5]。 在其他代谢病情况下,运动诱导的肝脏自噬已被证明对清除脂质和代"
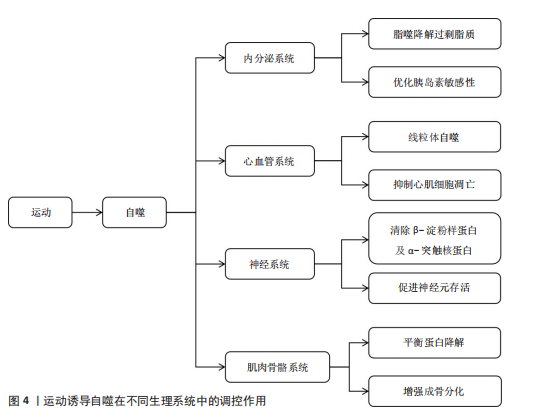

谢调控发挥关键作用[19]。运动诱导肝脏细胞自噬,将过量的三酰甘油和胆固醇降解,减轻内质网应激反应,改善肝脏脂肪变性[18]。除此之外,运动通过增加自噬相关基因1的表达启动自噬过程,并增强脂肪酸氧化能力。在脂肪肝患者中,自噬相关基因1不仅参与脂质调节,还参与介导对髓鞘形成和补体C1q/肿瘤坏死因子相关蛋白5等因子的活性调控[33]。进一步的研究发现,运动通过增加自噬相关蛋白7的表达促使自噬的延伸过程,缓解肥胖导致的内质网应激,显著改善肝脏的脂肪变性[34]。以上研究提示运动可从多种途径参与调控肝脏自噬,对改善肝脏代谢能力以及逆转肥胖导致的激素失衡等方面起到重要作用。 运动对机体内分泌系统的另一关键影响是通过腺苷酸活化蛋白激酶通路调控葡萄糖代谢及相关疾病的发生。腺苷酸活化蛋白激酶是细胞能量感应分子,在自噬调控、葡萄糖代谢中发挥重要的作用[35]。在糖尿病病理机制中,运动通过激活腺苷酸活化蛋白激酶刺激过氧化物酶体增殖物激活受体γ辅助活化因子α通路[36]、上调自噬相关蛋白Bcl2/腺病毒E1B相互作用蛋白3表达[37];此外,腺苷酸活化蛋白激酶激活后可通过PTEN诱导激酶1抑制内质网应激信号、调控肌醇需求酶1和PKR样内质网激酶/激活转录因子6的表达,进而恢复细胞稳态,减轻胰岛素抵抗并预防2型糖尿病的发生[18],说明运动诱导的自噬作用在糖尿病预防中具有潜在的作用,未来研究还需进一步探讨运动增强自噬作用以改善胰岛素敏感及糖尿病患者代谢。运动在慢性肾脏疾病中的治疗潜力同样值得关注,研究发现,运动可通过激活自噬减轻氧化应激、纤维化及炎症反应等,显著改善肾脏代谢葡萄糖,这一过程主要通过腺苷酸活化蛋白激酶通路诱导乙酰辅酶A羧化酶、自噬启动激酶1的磷酸化来增强自噬活性,进而缓解肾脏损伤[38]。以上研究从运动增强对内分泌调控角度展现了运动干预慢性肾脏病的应用前景。 总之,运动可通过激活自噬调控内分泌系统,自噬对内分泌系统的调控作用已逐渐成为代谢性疾病治疗的新靶点,自噬不仅可通过清除细胞内各种代谢废物和受损细胞器而维持内分泌系统稳态,还在多种代谢性疾病治疗中发挥关键作用。但不同运动类型如何在分子层面影响自噬活性及它们在不同内分泌疾病中起的作用尚不清晰。 2.3.2 运动诱导自噬在心血管系统中的调控作用 运动调控自噬在防治心血管疾病中的作用也得到学术界的关注[39]。自噬是细胞重要的降解途径之一,通过降解损伤的蛋白质、功能失调的细胞器和自身代谢产物维持细胞内环境的稳定,在参与心肌细胞适应性变化过程中发挥重要作用[10,40]。有研究表明,运动可借助自噬以减轻应激损伤,促进心肌功能恢复,从而保持心血管系统稳态[10,40]。 在应激引起的细胞能量失衡状态下,运动可以通过激活自噬以降解受损蛋白保持细胞能量稳态,减少心肌细胞凋亡,维持细胞内环境的平衡[9]。运动还能诱导线粒体自噬,选择性降解功能异常的线粒体,调控线粒体质量,从而发挥心脏保护作用[9]。有研究表明,在心肌缺血和缺氧损伤中,运动通过上调自噬相关基因1和微管相关蛋白1轻链3-Ⅱ等自噬标记基因表达促进受损细胞器与代谢产物的降解、增强线粒体腺苷三磷酸敏感性钾通道功能,减轻缺血损伤[14]。运动通过激活腺苷酸活化蛋白激酶-Unc-51样激酶1介导的自噬通路抑制病理性心脏肥大期心肌细胞凋亡、改善内质网应激,在防止不良心脏重塑的同时改善了心肌功能[41]。 在心力衰竭中,运动诱导的自噬能通过提高小鼠液泡分选蛋白34活性增强线粒体的氧化水平及腺苷三磷酸合成,有效减轻心脏负荷,表明运动是干预心力衰竭的有效方式[42]。在动脉粥样硬化的部分研究中,游泳等运动方式会通过上调微管相关蛋白轻链3与自噬相关基因1提高自噬水平,进而抑制可溶性的细胞间黏附分子1、基质金属蛋白酶9、白细胞介素6等炎症因子的释放,减少主动脉斑块的形成[15]。除此之外,运动训练还通过提升心肌的代谢作用、增进线粒体的生物合成,加快冠心病、心力衰竭及心脏移植术患者术后康复进程[43]。运动所带来的有益心血管健康作用,除了依靠分子机制外,还可通过提高心脏膜通透性增加心脏纤维及心壁厚度,提高心输出量及循环效率,从多角度维持心血管系统的健康[44]。 综上所述,运动诱发的自噬通过多途径在心脏的生理功能方面发挥重要调控作用,尤其在应激与疾病状态下,能够明显提高心肌能量代谢水平、减轻心肌细胞损伤以及修复受损的心肌组织等。然而,不同运动的种类、强度、频率如何精准调控自噬以及如何平衡运动负荷与自噬的调控作用仍需要进一步探究。因此,未来的研究重点应侧重于运动与自噬如何在心血管疾病发生发展的病理微环境中相互调控,为运动干预提供更具靶向性的理论支撑。 2.3.3 运动诱导自噬在神经系统中的调控作用 运动通过促进突触的形成、轴突的生长、神经可塑性和神经元再生等发挥显著的神经保护作用,改善认知能力、降低神经系统疾病发病风险[45]。运动触发的自噬,通过消除功能障碍的线粒体和积聚的有害蛋白来维持神经元内环境的稳态,改善神经元的线粒体功能、减少氧化应激损伤并维持神经元的完整性[7]。因此,运动可通过自噬途径预防神经退行性疾病。 运动诱导的自噬主要通过多个核心信号通路来实现,其中,沉默信息调节因子 1诱导的自噬通路和PTEN诱导激酶1-帕金蛋白通路依赖的线粒体自噬通路共同提高溶酶体活性,参与调节蛋白质量控制并改善线粒体动力学,直接抑制神经毒性聚集体的堆积,进而保护神经元[7]。同时,运动也可激活自噬相关基因1依赖的自噬通路,自噬相关基因1是调控细胞代谢的核心分子之一,与蛋白激酶B-叉头框蛋白O-雷帕霉素靶蛋白信号通路相互作用,维持蛋白质稳态和神经发生[16]。以上研究说明,运动不仅通过增强自噬促进神经元的存活,同时也通过调节神经元的代谢和蛋白质稳态来促进神经系统自我修复。 运动诱导的自噬在神经退行性疾病中起到重要的神经保护作用,例如在阿尔茨海默病及帕金森病中,运动通过自噬机制促进神经毒性蛋白(如α-突触核蛋白和β-淀粉样蛋白肽)的清除、减少错误折叠蛋白的积累[21,46]。自噬的激活可提高神经元的可塑性,延缓神经退行性病变过程,减少对神经元的损害[47]。有研究表明,运动诱导的自噬在改善神经退行性疾病的病理特征层面具有重要作用,例如在阿尔茨海默病中,运动诱导的自噬通过激活自噬相关基因1依赖性通路上调神经调控因子1表达,启动沉默信息调节因子 1-叉头框蛋白O-雷帕霉素靶蛋白信号级联反应,进而激活细胞外信号调节激酶-核糖体S6激酶-环腺苷酸反应元件结合蛋白通路,促进海马区神经发生和突触可塑性,这些机制不仅利于认知功能的恢复,还为改善阿尔茨海默病提供了一种新的方法[16]。但运动对这些自噬信号通路的时空调控机制以及运动诱导自噬中与其他神经保护途径的交互影响机制尚不清楚,后续研究有待进一步探讨运动诱导自噬在多种神经退行性疾病中的作用机制。在帕金森病中,运动通过激活自噬而减少多巴胺能神经元死亡、增强抗氧化防御功能、抑制氧化应激等,显著延缓神经退行性病变的病理进程;此外,通过运动增强神经可塑性与修复突触、轴突运输缺陷,显著改善学习记忆等[48],表明了运动可通过自噬途径促进神经系统恢复与功能重建。在缺血性脑损伤(缺血性脑血管病,如脑梗死)中,运动通过调节应激反应和血管生成协同增强自噬介导的神经保护作用[49]。有研究表明,早期损伤中的泛素结合蛋白上调诱导细胞外信号调节激酶的激活,自噬相关基因3通过减少内质网应激、激活磷酸肌醇3-激酶-沉默信息调节因子 1-雷帕霉素靶蛋白通路促进自噬活性,共同抑制神经元坏死、增强内源性防御机制、促进组织修复[50]。 综上所述,运动可通过介导自噬对神经系统发挥调控作用,既可以延缓神经退行性疾病的病理发展,也可以促进神经系统的修复及功能重塑。尽管现有研究初步阐明了许多可能的机制,然而不同种类运动对神经退行性疾病的作用机制尚需进一步阐明。 2.3.4 运动诱导自噬在肌肉骨骼系统中的调控作用 运动对维持肌肉骨骼健康至关重要,运动可以增大肌肉横截面积和肌肉强度,同时促进骨骼生成和骨骼成熟,从而有效降低肌肉骨骼损伤的发生风险[51]。在这一过程中,运动诱导的自噬是细胞内关键的降解和更新机制,运动诱导的自噬通过减少功能异常的线粒体、清除受损蛋白质和代谢废物促进线粒体再生,进而改善肌肉细胞的功能状态[52]。 运动诱导的自噬过程与一系列核心信号通路有关,尤其是与肌肉健康调控相关的自噬相关蛋白的激活,这些蛋白的激活不仅可以增强抗氧化防御系统,还会激活如骨形态生成蛋白等下游信号通路,改善骨骼肌无力和萎缩[53]。但是这些通路间具体的相互调控机制尚不完全清楚,未来还需要进一步阐明自噬相关分子在不同运动干预模式下的动态活性变化以及对骨骼肌健康的长期影响。运动诱导的自噬还能够调控骨骼肌生长和抗退化的多个核心信号通路[20],例如,激活磷酸肌醇3-激酶/沉默信息调节因子 1通路刺激抗凋亡蛋白的表达、抑制促凋亡因子的表达,抑制与肌肉萎缩有关的泛素连接酶及肌肉萎缩F-box蛋白,进而延缓肌肉衰退进程[13]。此外,运动通过抑制胰岛素样生长因子-雷帕霉素靶蛋白信号通路和核因子κB炎症通路抑制生长抑制因子的表达,从而刺激自噬相关基因1和自噬相关蛋白7等自噬相关分子的表达,并抑制由泛素-蛋白酶体系统介导的蛋白质降解,从而有效抵抗与衰老相关的肌肉退行性变[52]。 在骨骼健康调控中,运动通过诱导自噬相关复合物激活Wnt/β-连环蛋白信号通路,进而促进成骨分化及骨骼形成[54]。在成骨细胞中,自噬激活可清除错误折叠蛋白并抑制Ⅰ型胶原和Ⅱ型前胶原的异常蓄积,进而维持软骨细胞的稳态,这对于减轻关节炎等疾病引起的骨质流失和软骨退化至关重要;同时,运动通过腺苷酸活化蛋白激酶-雷帕霉素靶蛋白依赖的自噬激活核苷酸结合寡聚化结构域样受体蛋白3信号通路,促使炎症小泡的降解并降低焦磷酸盐毒性,进而维持软骨基质稳态[55]。以上机制证实了运动通过促进成骨、调控炎症反应影响骨骼和关节健康,进而延缓骨关节疾病的病理过程。运动对肌肉骨骼疾病的干预作用已在临床上得到验证,即运动可纠正肌肉萎缩、恢复肌肉功能并改善整体组织的健康状况[56],这凸显了运动作为肌肉骨骼系统再生与韧性调控因子的独特价值。但运动干预的最佳时机、强度及类型等问题还有待进一步界定。为了实现更好的临床干预,未来研究需进一步明确不同运动模式对机体自噬调控、骨骼肌肉健康及相关疾病防治的具体影响,尤其是在衰老和疾病状态下的作用机制。 综上所述,运动诱导的自噬在维持肌肉和骨骼的健康中起到重要作用,该过程涉及到复杂的分子机制和信号通路调控,未来有望阐释更多的运动和自噬相互作用机制,为改善老年及患病个体的肌肉骨骼功能提供新理论支持。 2.3.5 运动诱导自噬在不同生理状态下的作用 运动诱导自噬对不同生理系统的调控作用需重点关注运动方式、强度、频率及持续时间等因素,这些因素直接影响自噬的激活效率与调控效果,在不同生理状态下的作用差异尤为显著。运动对自噬的调控效应在健康个体与不同病理状态个体中可能存在明显差异[57]。 肌少症表现为肌肉量与肌力随年龄增长发生进行性下降,特别是在老年群体中更为常见[20]。研究表明,肌少症患者自噬活性低于健康个体,肌肉修复和重塑能力也因此降低[58]。长期进行有氧、抗阻运动可提升衰老骨骼肌中自噬相关蛋白的表达[59];然而,高强度运动会导致老年个体负荷过载[60],因此,需基于个体耐受能力动态调整运动强度与频率,后续需进一步围绕不同运动方式探索对肌少症的个体化方案。恶病质作为慢性病相关病理状态,通常表现有显著的消瘦及肌肉萎缩[61]。运动对恶病质的干预效果取决于类型-强度-病程阶段三要素[62],低中强度有氧联合抗阻训练能够通过自噬途径清除功能失调的细胞器与冗余蛋白,以此来改善肌肉质量和延缓萎缩,减轻代谢紊乱并增强修复能力[62-63],但是恶病质相关炎症、营养不良及代谢紊乱可能抑制自噬激活,故运动方案调整与执行还需结合患者具体的病理状态。 低氧条件下运动诱导的自噬作用和常规条件下不同[64],低氧能通过诱导Bcl2/腺病毒E1B相互作用蛋白3调节自噬相关基因的表达,以此影响细胞自噬过程[65]。另外,运动本身可通过调节胰岛素样生长因子1/雷帕霉素靶蛋白通路来增强线粒体自噬,清除受损线粒体,借此减轻氧化应激,维持细胞能量稳态。相关研究表明,腺苷酸活化蛋白激酶激活后能诱导线粒体自噬,并且会通过PTEN诱导激酶1-帕金蛋白通路非依赖途径激活TANK结合激酶1,推动线粒体裂变与自噬进程[66]。不同运动类型在低氧条件下的效应可能存在差别,基于此,考虑低氧环境与运动方式结合开展相关研究,可为临床及运动干预策略制定提供重要参考。此外,肥胖及代谢性疾病引发的脂肪堆积与代谢紊乱会对自噬正常功能产生干扰[67]。而运动可通过增强自噬活性促进脂肪酸氧化,减少肝脏和肌肉中的脂肪沉积,改善胰岛素敏感性与代谢功能。作为运动与胰岛素信号的关键分子,腺苷酸活化蛋白激酶能促进脂肪酸氧化、抑制糖原及蛋白质合成,有助于改善代谢状态[68]。 运动可通过激活腺苷酸活化蛋白激酶抑制雷帕霉素靶蛋白复合物1,以此引发自噬的激活,这一过程不仅可改善胰岛素敏感性,还有助于肌肉与肝脏细胞的修复。对于非酒精性脂肪肝患者来说,运动诱导自噬最终能够显著缓解肝脂肪变性,改善肝脏功能并减轻内质网应激[68-69]。研究发现,运动可激活自噬相关基因1所依赖的自噬通路,帮助改善神经元存活能力,从而推动突触可塑变化与神经细胞再生进程;自噬还在清除受损线粒体与错误折叠蛋白堆积过程中扮演着关键角色,这一重要作用对预防和治疗阿尔茨海默病、帕金森病等意义重大[70-71]。因此,不同模式运动在神经系统自噬调节过程中可能存在差异,基于此,需针对性制定个体化运动干预方案是研究重点。 综上所述,在不同生理状态下,运动对自噬的调控作用存在差异,并且这些差异和运动方式、强度、持续时间及病理特征都存在密切联系,未来研究应重点揭示运动如何在多样生理背景下对自噬的精准调控机制,以期为不同临床情况下的运动处方制定提供依据。 2.4 运动类型对自噬调控差异效应的剂量-反应关系 不同运动模式通过独特的分子通路激活自噬过程,直接影响自噬效率与持续时间。表2系统比较了耐力训练、抗阻训练、间歇训练在调控机制、信号通路及生理效应上的差异特征,揭示运动类型与自噬响应的特异性联系。 2.4.1 运动类型对自噬调控的基本差异 抗阻训练能够通过外部负荷刺激促进肌肉修复与增生,机械性的刺激作用于肌肉后可以调节肌肉蛋白质合成与降解平衡,提升肌肉质量与力量[72]。抗阻训练能够增强蛋白质合成过程、提升肌肉质量,调控肌纤维蛋白质合成与降解动态平衡[72-73]。抗阻训练在自噬调控中起到非常重要的作用,尤其是在肌肉修复与重塑的过程中能够激活腺苷酸活化蛋白激酶/雷帕霉素靶蛋白这一信号通路[74]。不仅如此,抗阻训练还可激活腺苷酸活化蛋白激酶途径,促使自噬启动激酶1磷酸化,诱导骨骼肌产生自噬流[58]。抗阻训练还能刺激外周血中单核细胞内的自噬,抑制核苷酸结合寡聚化结构域样受体蛋白3炎症小体激活、减少细胞凋亡,为肌肉细胞修复与功能恢复提供额外支持[75]。上述研究表明,抗阻训练通过调节自噬相关蛋白表达方式清除受损的细胞器及多余的蛋白质,从而维持肌肉健康、促进功能的恢复。 耐力训练可增强有氧代谢能力、改善心肺功能、增加能量供应。有氧运动可改善血脂状况,尤其是能够提高高密度脂蛋白胆固醇水平,对维护心脏健康具有重要作用[76]。无论是耐力型与力量型运动,长期训练都能够增强运动表现适应性,使身体更好地适应运动强度,维持最佳体能状态[77]。"


耐力训练通过显著激活线粒体自噬保障细胞能量供应[78],通过增强线粒体抗氧化及质控能力缓解氧化应激所带来的损伤[79],通过激活线粒体自噬提升细胞活性,进一步优化体内代谢平衡状态[80]。 高强度间歇训练因能够高效提升心血管功能、增加运动后代谢率以及优化氧气利用效率,被广泛认可为一种高效的训练方式[80]。有研究显示,同中等强度连续训练相比,高强度间歇训练改善最大摄氧量方面的效果更明显[80]。从细胞功能调控层面来看,高强度间歇训练可通过激活胰岛素样生长因子1/雷帕霉素靶蛋白信号轴增强细胞对能量应激的反应能力,触发自噬启动[81]。不仅如此,高强度间歇训练还通过提高自噬速率增强细胞的修复能力与对氧化应激的抵抗能力,对心血管、神经系统等产生积极影响[82]。 综上所述,3种运动方式在调控自噬方面展现出鲜明的特异性,分别在驱动肌肉修复与增生、优化线粒体质量和能量代谢和介导自噬反应方面有显著作用,表明运动类型、强度及持续时间调控自噬的作用机制是多样的。 2.4.2 不同运动方式对自噬激活效率的差异 低强度有氧运动维持细胞稳态并改善线粒体功能主要是通过持续较低的能量消耗激活自噬来完成的。研究表明,长期坚持有氧运动可减少心肌线粒体过度分裂与自噬,有利于改善心肌超微结构及功能[83]。高强度间歇训练通过激活胰岛素样生长因子1/雷帕霉素靶蛋白信号轴快速触发自噬反应,清除受损细胞器[84]。可见,低强度运动与高强度运动对自噬的调节具有差异:前者以温和的调节为主,后者可能通过更强的生理刺激触发自噬。 高强度运动可提升自噬相关蛋白的表达水平、增强细胞对能量应激的响应能力,从而推动自噬体形成,清除受损细胞器[85]。研究发现,相比持续中等强度运动,单次的高强度运动更能有效诱导自噬流,具体表现为两方面:提升微管相关蛋白1轻链3-Ⅱ/微管相关蛋白1轻链3-Ⅰ比值,降低泛素结合蛋白水平[85]。在低强度运动状态下,肌肉中自噬标记物的表达会随运动持续时间发生变化[86]。 短期训练与长期训练对自噬产生的效应存在差异。短期训练可快速启动自噬,提升细胞面对应激的应对能力,从而改善细胞代谢和线粒体功能[63],但要实现显著的细胞结构重塑较为困难,就自噬的长期适应效应而言,短期训练难以实现,尤其是训练强度偏低或运动量不足的情况[63]。长期训练可延长自噬反应时间,这对肌肉功能和其他组织稳态的维持是有利的[63]。值得注意的是,长期的有氧训练能够提高心肺功能、优化能量代谢效率并增强线粒体自噬,对于代谢健康的改善和细胞能量稳态维持具有重要作用[69];长期的有氧训练可激活腺苷酸活化蛋白激酶通路,增强肌肉的氧化能力,促进线粒体合成,最终改善肌肉功能[69]。抗阻训练可激活胰岛素样生长因子1/雷帕霉素靶蛋白信号轴,促进肌肉生长与重塑。研究证实,抗阻训练可提高肌肉的适应性与功能性,改善肌肉蛋白质的合成效率[87]。 因此,不同类型运动在自噬激活与自噬调节中存在差异。低强度运动以平缓的能量消耗帮助维持细胞稳态,促进线粒体功能的改善;高强度运动依靠剧烈生理刺激快速激活自噬。短期训练可快速激活自噬,改善代谢;长期训练可延长自噬反应时间,利于细胞对长期应激的适应。长期有氧训练与抗阻训练在代谢健康和肌肉功能方面展现出不同的优化路径。因此,阐明运动强度、周期及类型对自噬影响机制可为制定个体化的运动干预方案提供研究帮助。 2.4.3 不同生理状态下运动类型对自噬的调控差异 不同生理状态下,运动类型对自噬的调控效应会表现出一定的差异。针对健康个体,耐力训练主要通过对有氧代谢过程的强化来促进线粒体自噬,实现对心肺功能与细胞能量供给的改善。研究证实,运动是改善线粒体健康的有效方法,无论是肌肉组织或是其他机体组织都能获得有利帮助[88]。抗阻训练通过激活胰岛素样生长因子1/雷帕霉素靶蛋白信号轴促进肌肉的修复与再生,以此提升肌肉力量和质量[89]。腺苷酸活化蛋白激酶作为重要枢纽,能够在合成代谢与分解代谢途径发挥重要协调作用,以此平衡细胞及全身的能量供需[86]。抗阻训练与耐力训练能够有效保证骨骼肌健康,通过提升自噬活性清除衰老或受损的线粒体,帮助维持骨骼肌代谢稳态[88]。运动不仅能够强化自噬及线粒体自噬能力,还可增强细胞对氧化应激的抵抗力,改善线粒体功能,进而有效促进骨骼肌整体效能的优化[36,90]。 抗阻训练与有氧训练对自噬的调控作用在肌少症患者等特殊人群中具有明显不同。肌少症是因年龄增长或长期缺乏活动导致的肌肉量与力量进行性下降,因此需要选择适宜的运动类型调节自噬。抗阻训练能够通过增加肌肉负荷激活胰岛素样生长因子1/雷帕霉素靶蛋白信号通路,上调自噬相关蛋白的表达,从而促进肌肉的修复与生长[91],该训练有效促进肌肉增生的同时还可增强肌肉细胞的自噬功能,清除衰老及受损的细胞器[52,92]。 而耐力训练虽对改善代谢健康有一定帮助,但在促进肌肉修复与生长方面的效果不明显[52]。因此,对于肌少症患者群体而言,抗阻训练被视作更有效的干预方式[52]。 综上所述,运动类型对自噬的调节效应在不同生理状态下存在显著差异:在健康人群中,抗阻训练与耐力训练对自噬的调节各有侧重;而针对肌少症、恶病质等特殊人群,需选择正确合适的运动方式。未来研究需进一步探索不同生理状态下运动干预对自噬的精准调控机制,以期实现个体化运动处方的制定。"

| [1] MIZUSHIMA N, KOMATSU M. Autophagy: renovation of cells and tissues. Cell. 2011; 147(4):728-741. [2] LEVINE B, KROEMER G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176(1-2):11-42. [3] FRITZEN AM, MADSEN AB, KLEINERT M, et al. Regulation of autophagy in human skeletal muscle: effects of exercise, exercise training and insulin stimulation. J Physiol. 2016;594(3):745-761. [4] TIAN W, LI W, CHEN Y, et al. Phosphorylation of ULK1 by AMPK regulates translocation of ULK1 to mitochondria and mitophagy. FEBS Lett. 2015;589(15):1847-1854. [5] LIU X, NIU Y, YUAN H, et al. AMPK binds to Sestrins and mediates the effect of exercise to increase insulin-sensitivity through autophagy. Metabolism. 2015;64(6):658-665. [6] YOULE RJ, NARENDRA DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011; 12(1):9-14. [7] ZHAO N, XIA J, XU B. Physical exercise may exert its therapeutic influence on Alzheimer’s disease through the reversal of mitochondrial dysfunction via SIRT1-FOXO1/3-PINK1-Parkin-mediated mitophagy. J Sport Health Sci. 2021;10(1): 1-3. [8] ONODERA J, OHSUMI Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005; 280(36):31582-31586. [9] YAN L, VATNER DE, KIM SJ, et al. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005;102(39):13807-13812. [10] HE C, KLIONSKY DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67-93. [11] MIZUSHIMA N, YOSHIMORI T, OHSUMI Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011; 27:107-132. [12] KIM YC, GUAN KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125(1):25-32. [13] WHITE Z, TERRILL J, WHITE RB, et al. Voluntary resistance wheel exercise from mid-life prevents sarcopenia and increases markers of mitochondrial function and autophagy in muscles of old male and female C57BL/6J mice. Skelet Muscle. 2016; 6(1):45. [14] LI JY, PAN SS, WANG JY, et al. Changes in Autophagy Levels in Rat Myocardium During Exercise Preconditioning-Initiated Cardioprotective Effects. Int Heart J. 2019; 60(2):419-428. [15] LI Y, SUN D, ZHENG Y, et al. Swimming exercise activates aortic autophagy and limits atherosclerosis in ApoE-/- mice. Obes Res Clin Pract. 2020;14(3):264-270. [16] JANG Y. Endurance exercise-induced expression of autophagy-related protein coincides with anabolic expression and neurogenesis in the hippocampus of the mouse brain. Neuroreport. 2020;31(6):442-449. [17] WANG S, LI H, YUAN M, et al. Role of AMPK in autophagy. Front Physiol. 2022;13: 1015500. [18] CHENG F, DUN Y, CHENG J, et al. Exercise activates autophagy and regulates endoplasmic reticulum stress in muscle of high-fat diet mice to alleviate insulin resistance. Biochem Biophys Res Commun. 2022;601:45-51. [19] 魏元元,黄启超,许小君,等.运动干预非酒精性脂肪性肝病进程的研究进展[J].解放军医学杂志,2024,49(10):1207-1212. [20] 郭辉,孔健达,田春兰.线粒体自噬相关受体蛋白和信号通路在运动防治肌少症中的作用[J].中国组织工程研究,2024, 28(27):4397-4404. [21] 孔健达,解瑛傲,马雯,等.帕金森病相关线粒体功能障碍及运动对其潜在的改善作用[J].中国组织工程研究,2024, 28(27):4413-4420. [22] TAMARGO-GÓMEZ I, MARIÑO G. AMPK: Regulation of Metabolic Dynamics in the Context of Autophagy. Int J Mol Sci. 2018; 19(12):3812. [23] DI NARDO A, WERTZ MH, KWIATKOWSKI E, et al. Neuronal Tsc1/2 complex controls autophagy through AMPK-dependent regulation of ULK1. Hum Mol Genet. 2014; 23(14):3865-3874. [24] KOVALE L, SINGH MK, KIM J, et al. Role of Autophagy and AMPK in Cancer Stem Cells: Therapeutic Opportunities and Obstacles in Cancer. Int J Mol Sci. 2024;25(16):8647. [25] LEE JW, PARK S, TAKAHASHI Y, et al. The association of AMPK with ULK1 regulates autophagy. PLoS One. 2010;5(11):e15394. [26] LI J, QUAN C, HE YL, et al. Autophagy regulated by the HIF/REDD1/mTORC1 signaling is progressively increased during erythroid differentiation under hypoxia. Front Cell Dev Biol. 2022;10:896893. [27] 姚睿原,魏鸿渊,雷金叶,等.mTOR信号通路在疾病中的作用与调控机制[J].生命科学,2019,31(2):135-142. [28] SUN T, LI X, ZHANG P, et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6:7215. [29] 李建,曹炎,陈迎,等.自噬相关蛋白ATG5和ATG7在神经干细胞体外扩增中的作用[J].军事医学,2022,46(11):848-854. [30] KONG J, FAN R, ZHANG Y, et al. Oxidative stress in the brain-lung crosstalk: cellular and molecular perspectives. Front Aging Neurosci. 2024;16:1389454. [31] NARASIMHAN M, RAJASEKARAN NS. Exercise, Nrf2 and Antioxidant Signaling in Cardiac Aging. Front Physiol. 2016;7:241. [32] ZELKO IN, MARIANI TJ, FOLZ RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33(3):337-349. [33] CHUN SK, LEE S, YANG MJ, et al. Exercise-Induced Autophagy in Fatty Liver Disease. Exerc Sport Sci Rev. 2017;45(3):181-186. [34] HERRENBRUCK AR, BOLLINGER LM. Role of skeletal muscle autophagy in high-fat-diet-induced obesity and exercise. Nutr Rev. 2020;78(1):56-64. [35] 赵佳鹤,马欣雨,徐敬娅,等.自噬与非酒精性脂肪性肝病调控相关因子的关系[J].临床肝胆病杂志,2021,37(7):1713-1717. [36] 李兆进,郑鹏程,孔健达,等.基于不同组织和器官角度回顾PGC-1α在运动抗衰老中的作用[J].中国组织工程研究, 2024,28(29):4717-4725. [37] KIM D, SONG J, JIN EJ. BNIP3-Dependent Mitophagy via PGC1α Promotes Cartilage Degradation. Cells. 2021;10(7):1839. [38] JUSZCZAK F, VLASSEMBROUCK M, BOTTON O, et al. Delayed Exercise Training Improves Obesity-Induced Chronic Kidney Disease by Activating AMPK Pathway in High-Fat Diet-Fed Mice. Int J Mol Sci. 2020;22(1):350. [39] 陈志欣,张嘉敏,李素娟,等.运动调节巨噬细胞改善心血管疾病的分子机制研究进展[J].中国运动医学杂志,2024, 43(10):844-850. [40] 吴长勇,保苏丽,徐菲,等.运动调节自噬改善心血管疾病预后的研究进展[J].中国全科医学,2023,26(5):629-634. [41] MA Z, QI J, GAO L, et al. Role of Exercise on Alleviating Pressure Overload-Induced Left Ventricular Dysfunction and Remodeling via AMPK-Dependent Autophagy Activation. Int Heart J. 2020;61(5):1022-1033. [42] MACKENZIE MG, HAMILTON DL, MURRAY JT, et al. mVps34 is activated by an acute bout of resistance exercise. Biochem Soc Trans. 2007;35(Pt 5):1314-1316. [43] TAO L, BEI Y, LIN S, et al. Exercise Training Protects Against Acute Myocardial Infarction via Improving Myocardial Energy Metabolism and Mitochondrial Biogenesis. Cell Physiol Biochem. 2015;37(1):162-175. [44] FAYYAZ AI, DING Y. Mental stress, meditation, and yoga in cardiovascular and cerebrovascular diseases. Brain Circ. 2023;9(1):1-2. [45] TONG Y, DING Y, HAN Z, et al. Optimal rehabilitation strategies for early postacute stroke recovery: An ongoing inquiry. Brain Circ. 2023;9(4):201-204. [46] 夏杰,徐波.运动调节神经细胞自噬改善阿尔茨海默病[J].中国生物化学与分子生物学报,2020,36(7):748-755. [47] ROCCHI A, YAMAMOTO S, TING T, et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer’s disease. PLoS Genet. 2017; 13(8):e1006962. [48] JANG Y, KWON I, SONG W, et al. Endurance Exercise Mediates Neuroprotection Against MPTP-mediated Parkinson’s Disease via Enhanced Neurogenesis, Antioxidant Capacity, and Autophagy. Neuroscience. 2018;379:292-301. [49] DI RAIMONDO D, RIZZO G, MUSIARI G, et al. Role of Regular Physical Activity in Neuroprotection against Acute Ischemia. Int J Mol Sci. 2020;21(23):9086. [50] XING Y, YANG SD, WANG MM, et al. The beneficial roles of exercise training via autophagy in neurological diseases and possible mechanisms. Life Sci. 2019;221: 130-134. [51] KWON I, JANG Y, CHO JY, et al. Long-term resistance exercise-induced muscular hypertrophy is associated with autophagy modulation in rats. J Physiol Sci. 2018;68(3):269-280. [52] LIANG J, ZHANG H, ZENG Z, et al. Lifelong Aerobic Exercise Alleviates Sarcopenia by Activating Autophagy and Inhibiting Protein Degradation via the AMPK/PGC-1α Signaling Pathway. Metabolites. 2021;11(5):323. [53] SARTORI R, SCHIRWIS E, BLAAUW B, et al. BMP signaling controls muscle mass. Nat Genet. 2013;45(11):1309-1318. [54] CHEN X, SUN K, ZHAO S, et al. Irisin promotes osteogenic differentiation of bone marrow mesenchymal stem cells by activating autophagy via the Wnt//β-catenin signal pathway. Cytokine. 2020;136:155292. [55] LI Z, HUANG Z, ZHANG H, et al. Moderate-intensity exercise alleviates pyroptosis by promoting autophagy in osteoarthritis via the P2X7/AMPK/mTOR axis. Cell Death Discov. 2021;7(1):346. [56] MORA JC, VALENCIA WM. Exercise and Older Adults. Clin Geriatr Med. 2018;34(1): 145-162. [57] GRUMATI P, COLETTO L, SCHIAVINATO A, et al. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy. 2011;7(12):1415-1423. [58] PARK SS, SEO YK, KWON KS. Sarcopenia targeting with autophagy mechanism by exercise. BMB Rep. 2019;52(1):64-69. [59] WANG C, LIANG J, REN Y, et al. A Preclinical Systematic Review of the Effects of Chronic Exercise on Autophagy-Related Proteins in Aging Skeletal Muscle. Front Physiol. 2022;13:930185. [60] STERN G, PSYCHARAKIS SG, PHILLIPS SM. Effect of High-Intensity Interval Training on Functional Movement in Older Adults: A Systematic Review and Meta-analysis. Sports Med Open. 2023;9(1):5. [61] TISDALE MJ. Cachexia in cancer patients. Nat Rev Cancer. 2002;2(11):862-871. [62] DEVKOTA A, GAUTAM M, DHAKAL U, et al. The Interplay Between Physical Activity, Protein Consumption, and Sleep Quality in Muscle Protein Synthesis. J Physiol. 2025; 603(5):1121-1135. [63] IBATA N, TERENTJEV EM. Why exercise builds muscles: titin mechanosensing controls skeletal muscle growth under load. Biophys J. 2021;120(17):3649-3663. [64] 彭燕群,陈珍珍,翁锡全,等.低氧环境运动对营养性肥胖大鼠心肌细胞自噬相关蛋白表达的影响[J].中国病理生理杂志,2021,37(6):1027-1034. [65] BELLOT G, GARCIA-MEDINA R, GOUNON P, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29(10):2570-2581. [66] SEABRIGHT AP, FINE NHF, BARLOW JP, et al. AMPK activation induces mitophagy and promotes mitochondrial fission while activating TBK1 in a PINK1-Parkin independent manner. FASEB J. 2020;34(5): 6284-6301. [67] REUE K, WANG H. Mammalian lipin phosphatidic acid phosphatases in lipid synthesis and beyond: metabolic and inflammatory disorders. J Lipid Res. 2019; 60(4):728-733. [68] BABU AF, CSADER S, MÄNNISTÖ V, et al. Effects of exercise on NAFLD using non-targeted metabolomics in adipose tissue, plasma, urine, and stool. Sci Rep. 2022;12(1):6485. [69] VIOLLET B. The Energy Sensor AMPK: Adaptations to Exercise, Nutritional and Hormonal Signals//Spiegelman B, editor. Hormones, Metabolism and the Benefits of Exercise [Internet]. Cham (CH): Springer; 2017. [70] RIGHETTI E, ANTONELLO A, MARCHETTI L, et al. Mechanistic models of α-synuclein homeostasis for Parkinson’s disease: a blueprint for therapeutic intervention. Front Appl Math Stat. 2022;8:1060489. [71] THOMPSON TB, MEISL G, KNOWLES TPJ, et al. The role of clearance mechanisms in the kinetics of pathological protein aggregation involved in neurodegenerative diseases. J Chem Phys. 2021;154(12):125101. [72] WITARD OC, BANNOCK L, TIPTON KD. Making Sense of Muscle Protein Synthesis: A Focus on Muscle Growth During Resistance Training. Int J Sport Nutr Exerc Metab. 2022;32(1):49-61. [73] TIPTON KD, WOLFE RR. Exercise, protein metabolism, and muscle growth. Int J Sport Nutr Exerc Metab. 2001;11(1):109-132. [74] LUO L, LU AM, WANG Y, et al. Chronic resistance training activates autophagy and reduces apoptosis of muscle cells by modulating IGF-1 and its receptors, Akt/mTOR and Akt/FOXO3a signaling in aged rats. Exp Gerontol. 2013;48(4):427-436. [75] MEJÍAS-PEÑA Y, ESTÉBANEZ B, RODRIGUEZ-MIGUELEZ P, et al. Impact of resistance training on the autophagy-inflammation-apoptosis crosstalk in elderly subjects. Aging (Albany NY). 2017;9(2):408-418. [76] PATEL H, ALKHAWAM H, MADANIEH R, et al.Aerobic vs anaerobic exercise training effects on the cardiovascular system.. World J Cardiol. 2017;9(2):134-138. [77] HUGHES DC, ELLEFSEN S, BAAR K. Adaptations to Endurance and Strength Training. Cold Spring Harb Perspect Med. 2018;8(6):a029769. [78] YAN Z, LIRA VA, GREENE NP. Exercise training-induced regulation of mitochondrial quality. Exerc Sport Sci Rev. 2012;40(3):159-164. [79] WANG L, MASCHER H, PSILANDER N, et al. Resistance exercise enhances the molecular signaling of mitochondrial biogenesis induced by endurance exercise in human skeletal muscle. J Appl Physiol (1985). 2011; 111(5):1335-1344. [80] WU ZJ, WANG ZY, GAO HE, et al. Impact of high-intensity interval training on cardiorespiratory fitness, body composition, physical fitness, and metabolic parameters in older adults: A meta-analysis of randomized controlled trials. Exp Gerontol. 2021;150:111345. [81] HERZIG S, SHAW RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19(2):121-135. [82] MOHEBINEJAD M, KAZEMINASAB F, GHANBARI RAD M, et al. The Combined Effect of High-Intensity Interval Training and Time-Restricted Feeding on the AKT-IGF-1-mTOR Signaling Pathway in the Muscle Tissue of Type 2 Diabetic Rats. Nutrients. 2025;17(9):1404. [83] MA M, CHEN W, HUA Y, et al. Aerobic exercise ameliorates cardiac hypertrophy by regulating mitochondrial quality control and endoplasmic reticulum stress through M2 AChR. J Cell Physiol. 2021;236(9):6581-6596. [84] CARDINALE DA, GEJL KD, PETERSEN KG, et al. Short-term intensified training temporarily impairs mitochondrial respiratory capacity in elite endurance athletes. J Appl Physiol (1985). 2021;131(1):388-400. [85] ROCCHI A, HE C. Regulation of Exercise-Induced Autophagy in Skeletal Muscle. Curr Pathobiol Rep. 2017;5(2):177-186. [86] PINTO AP, DA ROCHA AL, MARAFON BB, et al. Impact of Different Physical Exercises on the Expression of Autophagy Markers in Mice. Int J Mol Sci. 2021;22(5):2635. [87] 夏志,徐飞,陈剑锋,等.mTOR信号转导在肌肉抗阻训练中的作用[J].生理科学进展,2009,40(4):375-377. [88] MEMME JM, ERLICH AT, PHUKAN G, et al. Exercise and mitochondrial health. J Physiol. 2021;599(3):803-817. [89] KJØBSTED R, HINGST JR, FENTZ J, et al. AMPK in skeletal muscle function and metabolism. FASEB J. 2018;32(4):1741-1777. [90] VAINSHTEIN A, TRYON LD, PAULY M, et al. Role of PGC-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle. Am J Physiol Cell Physiol. 2015;308(9):C710-C719. [91] CAO Y, ZHOU J, QUAN H, et al. Resistance training alleviates muscle atrophy and muscle dysfunction by reducing inflammation and regulating compromised autophagy in aged skeletal muscle. Front Immunol. 2025;16:1597222. [92] FAN J, KOU X, JIA S, et al. Autophagy as a Potential Target for Sarcopenia. J Cell Physiol. 2016;231(7):1450-1459. |
| [1] |
Dong Chunyang, Zhou Tianen, Mo Mengxue, Lyu Wenquan, Gao Ming, Zhu Ruikai, Gao Zhiwei.
Action mechanism of metformin combined with Eomecon chionantha Hance dressing in treatment of deep second-degree burn wounds#br#
#br#
[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(8): 2001-2013.
|
| [2] | Yang Xuetao, Zhu Menghan, Zhang Chenxi, Sun Yimin, Ye Ling. Applications and limitations of antioxidant nanomaterials in oral cavity [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(8): 2044-2053. |
| [3] | Liu Anting, Lu Jiangtao, Zhang Wenjie, He Ling, Tang Zongsheng, Chen Xiaoling. Regulation of AMP-activated protein kinase by platelet lysate inhibits cadmium-induced neuronal apoptosis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1800-1807. |
| [4] | Sun Yaotian, Xu Kai, Wang Peiyun. Potential mechanisms by which exercise regulates iron metabolism in immune inflammatory diseases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1486-1498. |
| [5] | You Huijuan, Wu Shuzhen, Rong Rong, Chen Liyuan, Zhao Yuqing, Wang Qinglu, Ou Xiaowei, Yang Fengying. Macrophage autophagy in lung diseases: two-sided effects [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1516-1526. |
| [6] | Jia Jinwen, Airefate·Ainiwaer, Zhang Juan. Effects of EP300 on autophagy and apoptosis related to allergic rhinitis in rats [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1439-1449. |
| [7] | Liu Kexin, , Hao Kaimin, Zhuang Wenyue, , Li Zhengyi. Autophagy-related gene expression in pulmonary fibrosis models: bioinformatic analysis and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1129-1138. |
| [8] | Hu Jing, Zhu Ling, Xie Juan, Kong Deying, Liu Doudou. Autophagy regulates early embryonic development in mice via affecting H3K4me3 modification [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1147-1155. |
| [9] | Wen Fan, Xiang Yang, Zhu Huan, Tuo Yanfang, Li Feng. Exercise improves microvascular function in patients with type 2 diabetes [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1225-1235. |
| [10] | Yang Xiao, Bai Yuehui, Zhao Tiantian, Wang Donghao, Zhao Chen, Yuan Shuo. Cartilage degeneration in temporomandibular joint osteoarthritis: mechanisms and regenerative challenges [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 926-935. |
| [11] | Dong Chao, Zhao Mohan, Liu Yunan, Yang Zeli, Chen Leqin, Wang Lanfang. Effects of magnetic nano-drug carriers on exercise-induced muscle injury and inflammatory response in rats [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(2): 345-353. |
| [12] | Zheng Wen, Zhu Dongsheng, Wang Xiaodong. Secreted modular calcium binding protein regulates autophagy in the acetabular cartilage of rats with developmental dysplasia of the hip [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(18): 4618-4626. |
| [13] | Liao Xingzhuan, Li Guangdi, Wu Yabin, Liu Xingyu, Wan Jiajia. Molecular mechanisms underlying non-coding RNA regulation of ferroptosis in osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(18): 4713-4725. |
| [14] | Yang Jiangxi, Li Huangyan, Zhang Yeting, Yu Zuoyin . Research hotspots and thematic evolution in the field of exercise interventions for multiple sclerosis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(18): 4771-4781. |
| [15] | Ji Dejiang, Zhang Xiaojing, Ye Gaxi. Role and mechanism by which acupuncture regulates autophagy in a rat model of cerebral hemorrhage [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(16): 4067-4076. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||