Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (5): 1129-1138.doi: 10.12307/2026.040
Previous Articles Next Articles
Autophagy-related gene expression in pulmonary fibrosis models: bioinformatic analysis and experimental validation
Liu Kexin1, 2, Hao Kaimin2, Zhuang Wenyue2, 3, Li Zhengyi1
- 1Laboratory Academy, Jilin Medical University, Jilin 132001, Jilin Province, China; 2Beihua University, Jilin 132001, Jilin Province, China; 3Zhejiang Wanli University, Ningbo 315100, Zhejiang Province, China
-
Received:2024-11-19Accepted:2025-01-24Online:2026-02-18Published:2025-06-23 -
Contact:Li Zhengyi, PhD, Professor, Master’s supervisor, Laboratory Academy, Jilin Medical University, Jilin 132001, Jilin Province, China -
About author:Liu Kexin, MS candidate, Laboratory Academy, Jilin Medical University, Jilin 132001, Jilin Province, China; Beihua University, Jilin 132001, Jilin Province, China -
Supported by:Jilin Province Scientific and Technology Development Project, No. YDZJ202201ZYTS637 (to ZWY); Jilin Medical University Graduate Innovation Program, No. 2023yy01 (to LKX)
CLC Number:
Cite this article
Liu Kexin, , Hao Kaimin, Zhuang Wenyue, , Li Zhengyi. Autophagy-related gene expression in pulmonary fibrosis models: bioinformatic analysis and experimental validation[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1129-1138.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
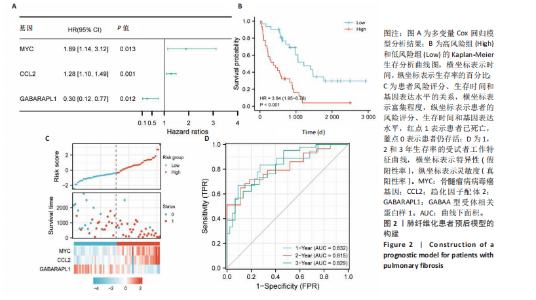
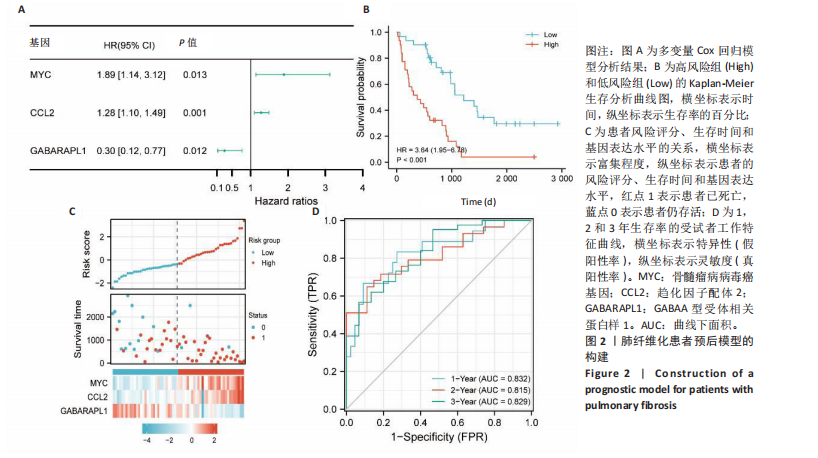
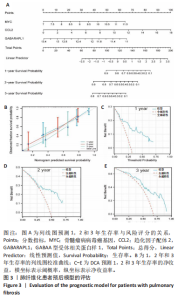
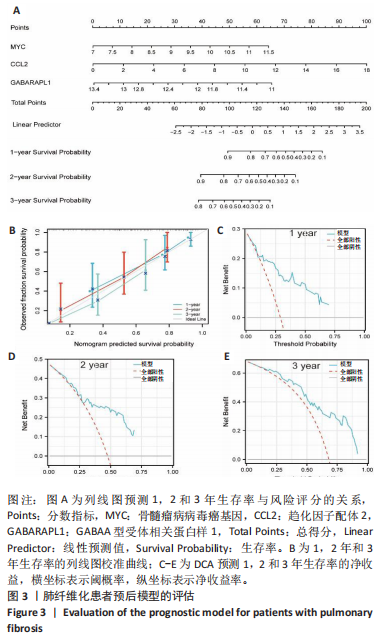
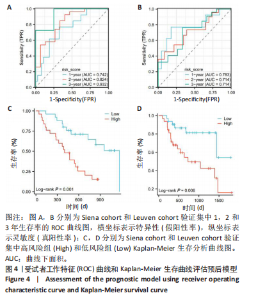
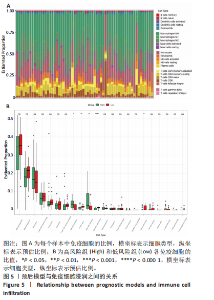
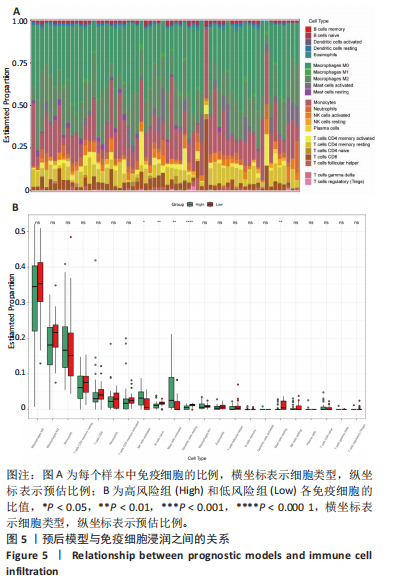
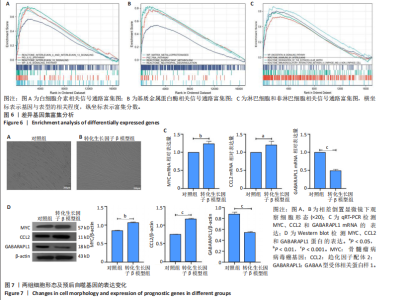
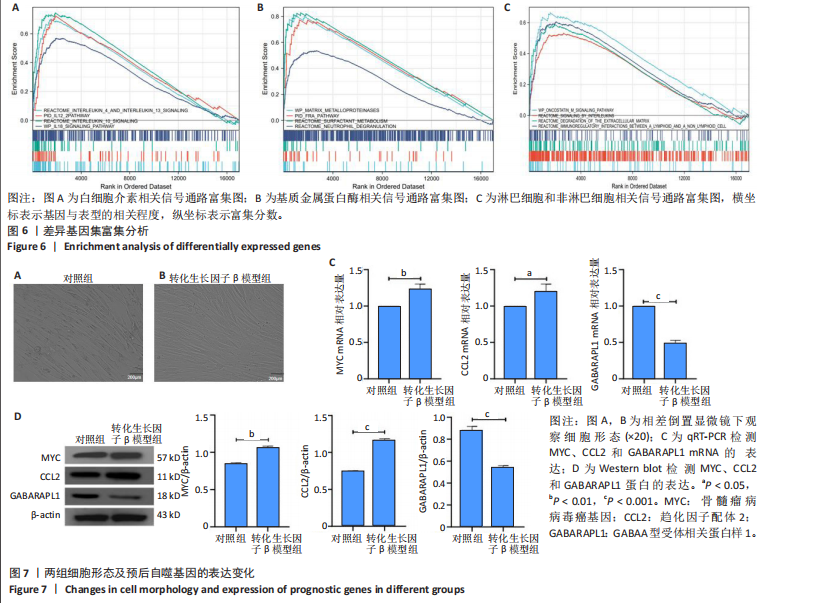
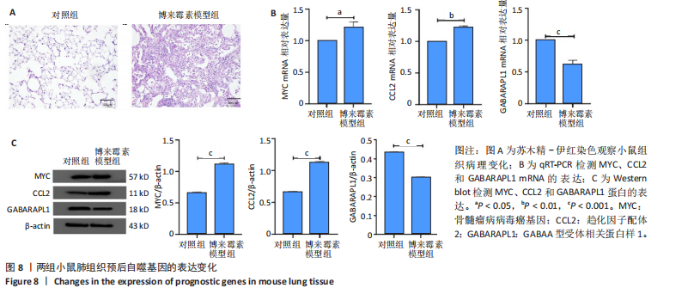
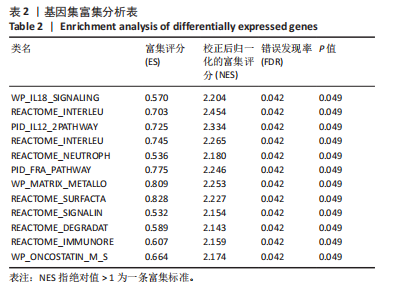
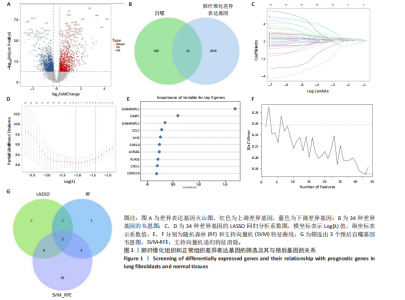
2.1 差异表达基因的筛选及其与预后基因的关联 在数据集GSE70866中鉴定了来自Freiburg cohort的20名健康供者和62例肺纤维化患者的差异表达基因(其中高风险组41个样本,低风险组21个样本),包括1 099个上调基因和1 551个下调基因(图1A),鉴定出34个自噬相关的差异表达基因(图1B)。在LASSO回归模型中选择了9个候选基因(图1C,D);在随机森林(random forest,RF)模型中鉴定出9个候选基因(图1E);在支持向量机(support vector machine,SVM)模型中选择了33个候选基因(图1F)。基于上述3种模型筛选3个预后自噬基因(MYC、CCL2、GABARAPL1)(图1G)。 2.2 预后模型的构建 使用多因素Cox回归分析3个影响肺纤维化的独立因素,其中CCL2和MYC为危险因素,GABARAPL1为保护因素。根据这3个基因得到肺纤维化患者预后的风险评分=0.64×MYC exp+0.25×CCL2 exp+(-1.21×GABARAPL1 exp)的中位数(图2A),将肺纤维化样本根据风险评分分为高风险组和低风险组。K-M生存曲线显示(图2B),较低的风险评分与较高的生存率相关。患者的风险评分、生存时间和基因表达水平有关(图2C)。ROC曲线用于评价风险评分预测患者的预后能力,并计算1,2和3年的曲线下面积(area under curve,AUC)分别为0.832,0.815和0.829(图2D)。 2.3 预后模型的评估 通过计算肺纤维化患者在诊断后1,2和3年的生存率,构建列线图来估计肺纤维化患者的预后(图3A),并应用Siena cohort和Leuven cohort验证集进行验证。C指数、校准曲线(图3B)和决策曲线(图3C-E)分析表明,列线图模型的精度高[0.753(0.714-0.792)]。在Siena cohort验证集中,AUC值分别为0.742,0.824和0.932(图4A)。在Leuven cohort验证集中,AUC值分别为0.783、0.714和0.714(图4B)。Siena cohort(图4C)和Leuven cohort验证集的Kaplan-Meier生存分析曲线显示(图4D),高风险组的存活率明显比低风险组低。 2.4 免疫细胞浸润 条形图显示了每个样品中免疫细胞的比例(图5A)。低风险组中活化的肥大细胞(P < 0.001)和活化的自然杀伤细胞(P < 0.01)浸润更多;高风险组中活化的CD4记忆T细胞(P < 0.01)、幼稚B细胞(P < 0.001)、静息下的树突状细胞(P < 0.001)和静息的肥大细胞(P < 0.001)浸润更多(图5B)。 2.5 基因集富集分析 为了确定2个风险组的功能,对差异基因进行了基因集富集分析。共发现87个基因集[错误发现率(FDR) < 0.25和P < 0.05],包括白细胞介素相关(白细胞介素4,白细胞介素10,白细胞介素12,白细胞介素13)途径,基质金属蛋白酶、淋巴细胞和非淋巴细胞之间的相互调节作用(图6)。曲线表示富集评分(enrichment score,ES)的情况,结果显示校正后归一化的ES(NES)值为正,峰出现在左侧,基因集中核心分子主要集中在左侧高风险组中。基因集富集分析的详细信息如表2所示。 2.6 体外细胞模型中自噬基因的表达 2.6.1 体外模型建立 显微镜下观察HFL-1细胞形态发现,对照组HFL-1细胞呈梭形且边界清晰,排列"
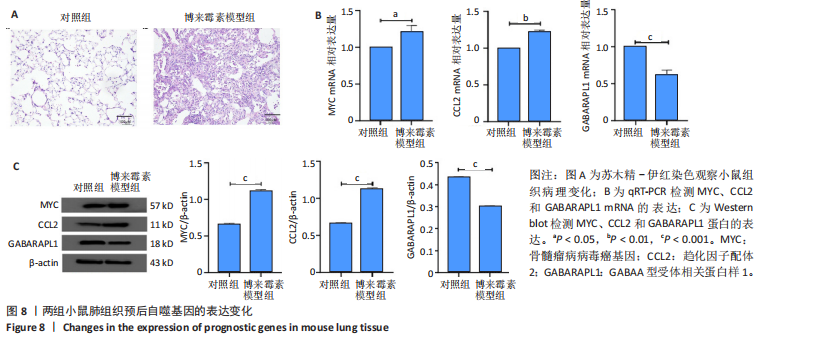
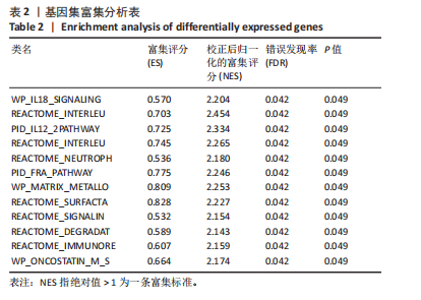
规则,模型组细胞间隙增宽,HFL-1细胞呈扁平状,边界不清晰,细胞核比例增大,并且转化生长因子β1刺激后,HFL-1细胞间的连接变得更加疏松,以上形态学改变证明细胞造模成功(图7A,B)。 2.6.2 qRT-PCR检测 结果显示,与对照组比,转化生长因子β1模型组HFL-1细胞的MYC和CCL2 mRNA表达水平升高(P < 0.01,P < 0.05),GABARAPL1 mRNA表达水平降低(P < 0.001,图7C)。 2.6.3 Western blot检测 结果显示,与对照组相比,转化生长因子β1模型组HFL-1细胞的MYC和CCL2 蛋白表达水平升高(P < 0.01,P < 0.001),GABARAPL1 蛋白表达水平降低(P < 0.001,图7D)。 2.7 体内动物模型中自噬基因的表达 2.7.1 实验动物数量分析 实验选用小鼠共20只,分为2组,每组10只,在造模过程中各组小鼠的死亡数为0。 2.7.2 小鼠肺组织苏木精-伊红染色结果 对照组小鼠肺组织肺泡结构清晰完整,未发现肺泡炎性渗出及纤维化病变;而博莱霉素模型组肺泡结构紊乱,肺泡壁显著增厚,炎性细胞显著浸润,提示肺纤维化小鼠建模成功(图8A)。 2.7.3 qRT-PCR检测结果 与对照组相比,博莱霉素模型组小鼠肺组织中的MYC和CCL2 mRNA表达水平明显升高(P < 0.05,P < 0.01),而GABARAPL1 mRNA表达水平则有所下降(P < 0.001,图8B)。 2.7.4 Western blot检测结果 与对照组相比,博莱霉素模型组小鼠肺组织中的MYC和CCL2 蛋白表达水平明显升高(P < 0.001),而GABARAPL1 蛋白表达水平则降低(P < 0.001,图8C)。"
| [1] CHANDA D, OTOUPALOVA E, SMITH SR, et al. Developmental pathways in the pathogenesis of lung fibrosis. Mol Aspects Med. 2019;65:56-69. [2] LI X, WANG Y, LIANG J, et al. Bergenin attenuates bleomycin‐induced pulmonary fibrosis in mice via inhibiting TGF-β1signaling pathway. Phytother Res. 2021;35(10):5808-5822. [3] GAO Q, CHANG X, YANG M, et al. LncRNA MEG3 restrained pulmonary fibrosis induced by NiO NPs via regulating hedgehog signaling pathway-mediated autophagy.Environ Toxicol. 2022;37(1):79-91. [4] YAO Y, JIANG P, CHAO B, et al. GIMAP6 regulates autophagy, immune competence, and inflammation in mice and humans. J Exp Med. 2022; 219(6):e20201405. [5] HEUKELS P, MOOR CC, VON DER THÜSEN JH, et al. Inflammation and immunity in IPF pathogenesis and treatment. Respir Med. 2019;147:79-91. [6] ZHENG D, GUO J, LIANG Z, et al. Supramolecular Nanofibers Ameliorate Bleomycin‐Induced Pulmonary Fibrosis by Restoring Autophagy. Adv Sci (Weinh). 2024;11(28):e2401327. [7] WANG B, WANG Y, ZHANG J, et al. ROS-induced lipid peroxidation modulates cell death outcome: mechanisms behind apoptosis, autophagy, and ferroptosis. Arch Toxicol. 2023;97(6):1439-1451. [8] LEE C, KWAK S H, HAN J, et al. Repositioning of ezetimibe for the treatment of idiopathic pulmonary fibrosis. Eur Respir J. 2024;63(5):2300580. [9] DUAN JX, GUAN XX, YANG HH, et al. Vasoactive intestinal peptide attenuates bleomycin-induced murine pulmonary fibrosis by inhibiting epithelial-mesenchymal transition: restoring autophagy in alveolar epithelial cells. Int Immunopharmacol. 2021;101(Pt B):108211. [10] TANG Q, XING C, LI M, et al. Pirfenidone ameliorates pulmonary inflammation and fibrosis in a rat silicosis model by inhibiting macrophage polarization and JAK2/STAT3 signaling pathways. Ecotoxicol Environ Saf. 2022;244:114066. [11] ZHOU Z, JIANG X, LIU D, et al. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5(3):321-328. [12] REED M, MORRIS SH, OWCZARCZYK AB, et al. Deficiency of autophagy protein Map1-LC3b mediates IL-17-dependent lung pathology during respiratory viral infection via ER stress-associated IL-1. Mucosal Immunol. 2015;8(5):1118-1130. [13] HE Y, THUMMURI D, ZHENG G, et al. Cellular senescence and radiation-induced pulmonary fibrosis. Transl Res. 2019;209:14-21. [14] LI S, LI C, PANG X, et al. Metformin attenuates silica-induced pulmonary fibrosis by activating autophagy via the AMPK-mTOR signaling pathway. Front Pharmacol. 2021;12:719589. [15] LI X, HUANG K, LIU X, et al. Ellagic acid attenuates BLM-induced pulmonary fibrosis via inhibiting Wnt signaling pathway. Front Pharmacol. 2021;12:639574. [16] TZOUVELEKIS A, GOMATOU G, BOUROS E, et al. Common pathogenic mechanisms between idiopathic pulmonary fibrosis and lung cancer. Chest. 2019;156(2):383-391. [17] MU E, WANG J, CHEN L, et al. Ketogenic diet induces autophagy to alleviate bleomycin-induced pulmonary fibrosis in murine models. Exp Lung Res. 2021;47(1):26-36. [18] WYNN TA. Integrating mechanisms of pulmonary fibrosis. J Exp Med. 2011;208(7):1339-1350. [19] FU W, FANG X, WU L, et al. Neogambogic acid relieves myocardial injury induced by sepsis via p38 MAPK/NF-κB pathway. Korean J Physiol Pharmacol. 2022;26(6):511-518. [20] ZHAO J, WU Q, YANG T, et al. Gaseous signal molecule SO2 regulates autophagy through PI3K/AKT pathway inhibits cardiomyocyte apoptosis and improves myocardial fibrosis in rats with type II diabetes. Korean J Physiol Pharmacol. 2022;26(6):541-556. [21] 王晓旭. 羟基红花黄色素A通过自噬溶酶体途径缓解特发性肺纤维化机制研究[D].保定:河北大学,2024. [22] 菅若男,甄华,郭晶晶.基于PI3K/AKT/mTOR信号通路激活自噬通路研究荜茇改善小鼠肺纤维化的机制[J].临床肺科杂志,2024, 29(5):675-682. [23] 李鹏,牛璐,赵国静,等.基于自噬探究补肺活血胶囊对博莱霉素诱发小鼠肺纤维化的抑制作用[J].中成药杂志,2024,46(4):1337-1342. [24] MENG Y, PAN M, ZHENG B, et al. Autophagy attenuates angiotensin II-induced pulmonary fibrosis by inhibiting redox imbalance-mediated NOD-like receptor family pyrin domain containing 3 inflammasome activation. Antioxid Redox Signal. 2019;30(4):520-541. [25] SHANG C, HASSAN B, HAQUE M, et al. Crizotinib resistance mediated by autophagy is higher in the stem-like cell subset in ALK-positive anaplastic large cell lymphoma, and this effect is MYC-dependent. Cancers (Basel). 2021;13(2):181. [26] LIU S, YANG Q, DONG B, et al. Gypenosides Attenuate Pulmonary Fibrosis by Inhibiting the AKT/mTOR/c-Myc Pathway. Front Pharmacol. 2022;12:806312. [27] 阚泉,张岩,王海涛,等.抗氧化剂超氧化物歧化酶对大鼠肺巨噬细胞二氧化硅条件上清介导的肺成纤维细胞增殖及c-myc表达的影响[J].中国组织工程研究,2022,26(14):2202-2206. [28] JIN Y, QIU J, LU X, et al. C-MYC inhibited ferroptosis and promoted immune evasion in ovarian cancer cells through NCOA4 mediated ferritin autophagy. Cells. 2022;11(24):4127. [29] ENTRIALGO-CADIERNO R, CUETO-UREÑA C, WELCH C, et al. The phospholipid transporter PITPNC1 links KRAS to MYC to prevent autophagy in lung and pancreatic cancer. Mol Cancer. 2023;22(1):86. [30] LIN HJ, LIU HH, LIN CD, et al. Cytolethal distending toxin enhances radiosensitivity in prostate cancer cells by regulating autophagy. Front Cell Infect Microbiol. 2017;7:223. [31] KANMANI P, KIM H. Probiotics counteract the expression of hepatic profibrotic genes via the attenuation of TGF-β/SMAD signaling and autophagy in hepatic stellate cells. PLoS One. 2022;17(1):e0262767. [32] DENG X, MERCER PF, SCOTTON CJ, et al. Thrombin induces fibroblast CCL2/JE production and release via coupling of PAR1 to Gαq and cooperation between ERK1/2 and Rho kinase signaling pathways. Mol Biol Cell. 2008;19(6):2520-2533. [33] XU H, WANG J, AL‐NUSAIF M, et al. CCL2 promotes metastasis and epithelial–mesenchymal transition of non‐small cell lung cancer via PI3K/Akt/mTOR and autophagy pathways. Cell Prolif. 2024;57(3): e13560. [34] GAO Y, CHEN S, JIAO S, et al. ATG5-regulated CCL2/MCP-1 production in myeloid cells selectively modulates anti-malarial CD4+ Th1 responses. Autophagy. 2024;20(6):1398-1417. [35] NIINIKOSKI I, KOUKI S, KOHO N, et al. Evaluation of VEGF-A and CCL2 in dogs with brachycephalic obstructive airway syndrome or canine idiopathic pulmonary fibrosis and in normocephalic dogs. Res Vet Sci. 2022;152:557-563. [36] LU G, RAO D, ZHOU M, et al. Autophagic Network Analysis of the Dual Effect of Sevoflurane on Neurons Associated with GABARAPL1 and 2. Biomed Res Int. 2020;2020:1587214. [37] KEULERS TG, LIBREGTS SF, BEAUMONT JEJ, et al. Secretion of proangiogenic extracellular vesicles during hypoxia is dependent on the autophagy-related protein GABARAPL1.J Extracell Vesicles. 2021;10(14):e12166. [38] MENG J, XU L, MA B, et al. GABARAPL1 is essential for ACR-induced autophagic cell death of mouse Leydig cells. Ecotoxicol Environ Saf. 2025;289:117426. |
| [1] | Hu Jing, Zhu Ling, Xie Juan, Kong Deying, Liu Doudou. Autophagy regulates early embryonic development in mice via affecting H3K4me3 modification [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1147-1155. |
| [2] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [3] | Liu Lin, Liu Shixuan, Lu Xinyue, Wang Kan. Metabolomic analysis of urine in a rat model of chronic myofascial trigger points [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1585-1592. |
| [4] | Li Huayuan, Li Chun, Liu Junwei, Wang Ting, Li Long, Wu Yongli. Effect of warm acupuncture on PINK1/Parkin pathway in the skeletal muscle of rats with chronic fatigue syndrome [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1618-1625. |
| [5] | Zhou Panpan, Cui Yinglin, Zhang Wentao, Wang Shurui, Chen Jiahui, Yang Tong . Role of cellular autophagy in cerebral ischemic injury and the regulatory mechanism of traditional Chinese medicine [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1650-1658. |
| [6] | Zhu Hanmin, Wang Song, Xiao Wenlin, Zhang Wenjing, Zhou Xi, He Ye, Li Wei, . Mitophagy regulates bone metabolism [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1676-1683. |
| [7] | Zhao Jiacheng, Ren Shiqi, Zhu Qin, Liu Jiajia, Zhu Xiang, Yang Yang. Bioinformatics analysis of potential biomarkers for primary osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1741-1750. |
| [8] | Zhang Zhenyu, Liang Qiujian, Yang Jun, Wei Xiangyu, Jiang Jie, Huang Linke, Tan Zhen. Target of neohesperidin in treatment of osteoporosis and its effect on osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1437-1447. |
| [9] | Zhang Haojun, Li Hongyi, Zhang Hui, Chen Haoran, Zhang Lizhong, Geng Jie, Hou Chuandong, Yu Qi, He Peifeng, Jia Jinpeng, Lu Xuechun. Identification and drug sensitivity analysis of key molecular markers in mesenchymal cell-derived osteosarcoma [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1448-1456. |
| [10] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [11] | Zheng Rongfa, Mo Weibin, Huang Peng, Chen Junji, Liang Ting, Zi Fangyu, Li Guofeng. Effects of electroacupuncture on the expression of metabolic enzymes and autophagy genes in gastrocnemius muscle tissues of exercising rats [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1127-1136. |
| [12] | Chen Yuning, Jiang Ying, Liao Xiangyu, Chen Qiongjun, Xiong Liang, Liu Yue, Liu Tong. Buqi Huoxue Compounds intervene with the expression of related factors and autophagy related proteins in a rat model of cerebral ischemia/reperfusion [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1152-1158. |
| [13] | Liu Lingyun, He Guixin, Qin Weibin, Song Hui, Zhang Liwen, Tang Weizhi, Yang Feifei, Zhu Ziyi, Ou Yangbin . Improvement of myocardial injury by traditional Chinese medicine: mitochondrial calcium homeostasis mediates macrophage autophagy and pyroptosis pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1276-1284. |
| [14] | Xu Tianjie, Fan Jiaxin, Guo Xiaoling, Jia Xiang, Zhao Xingwang, Liu kainan, Wang Qian. Metformin exerts a protective effect on articular cartilage in osteoarthritis rats by inhibiting the PI3K/AKT/mTOR pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1003-1012. |
| [15] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||