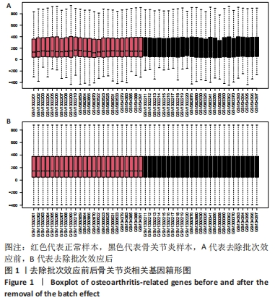
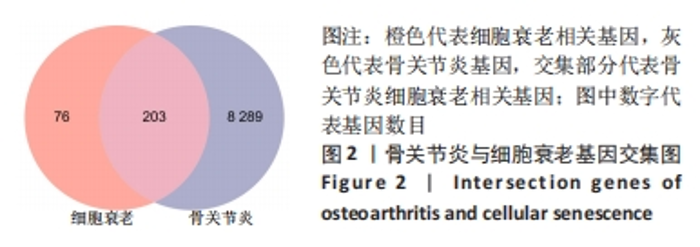
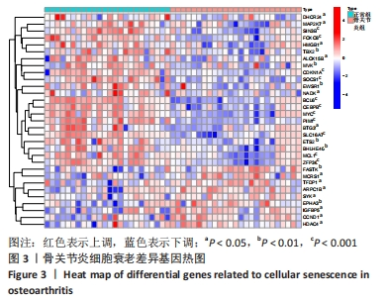
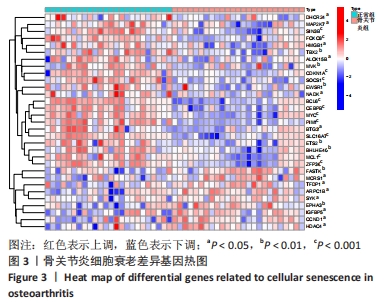
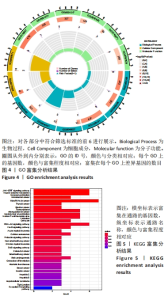
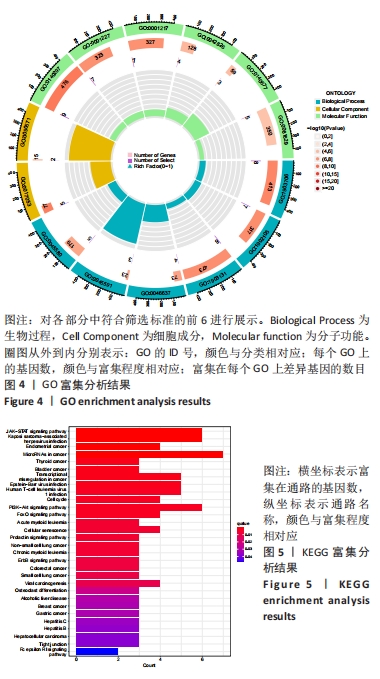
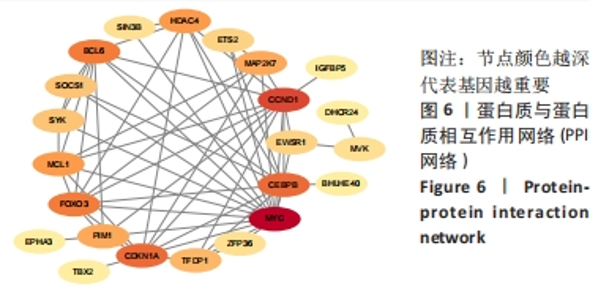
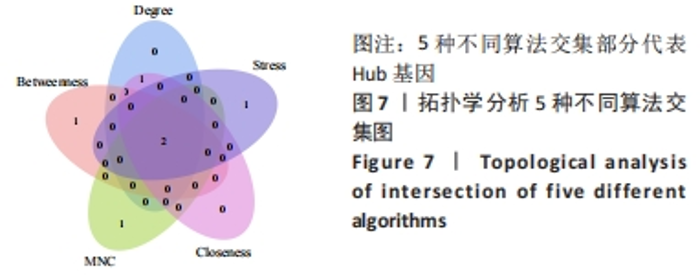
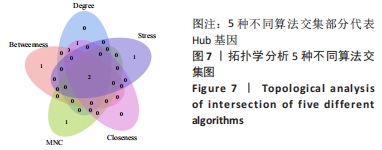
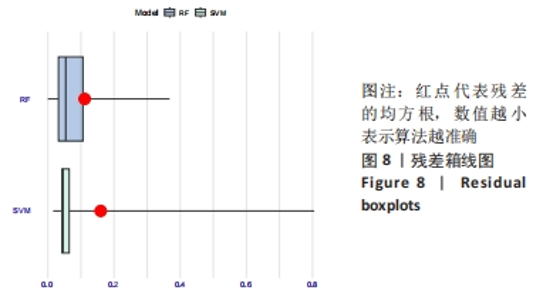
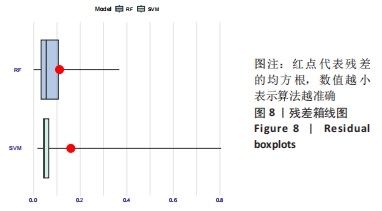
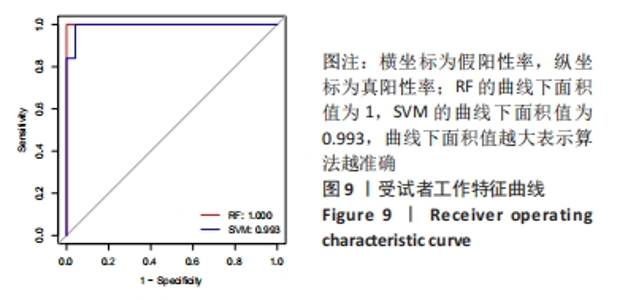
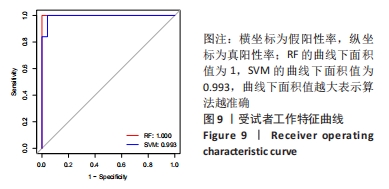
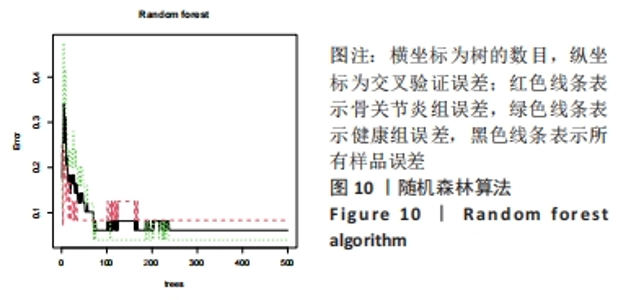
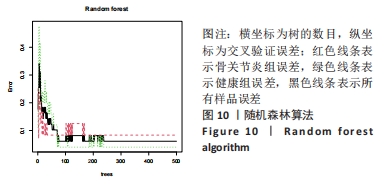
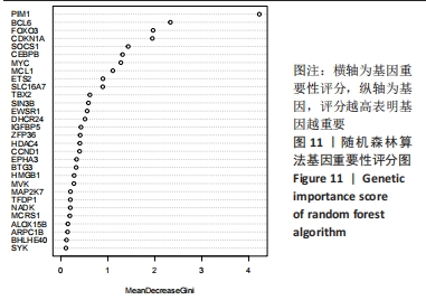
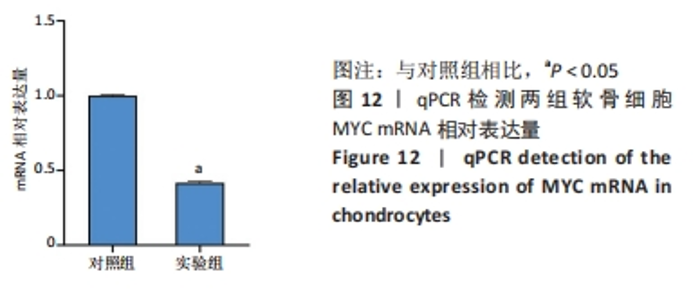
Chinese Journal of Tissue Engineering Research ›› 2024, Vol. 28 ›› Issue (20): 3196-3202.doi: 10.12307/2024.344
Previous Articles Next Articles
Machine learning combined with bioinformatics to identify and validate key genes for cellular senescence in osteoarthritis
Yuan Changshen1, Liao Shuning2, Li Zhe2, Wu Siping2, Chen Lewei2, Liu Jinyi2, Li Yanhong2, Duan Kan1
- 1Orthopedic Department of the Limbs, The First Affiliated Hospital of Guangxi University of Chinese Medicine, Nanning 530023, Guangxi Zhuang Autonomous Region, China; 2School of Postgraduate, Guangxi University of Chinese Medicine, Nanning 530000, Guangxi Zhuang Autonomous Region, China
-
Received:2023-04-07Accepted:2023-06-10Online:2024-07-18Published:2023-09-11 -
Contact:Duan Kan, MD, Chief physician, Orthopedic Department of the Limbs, The First Affiliated Hospital of Guangxi University of Chinese Medicine, Nanning 530023, Guangxi Zhuang Autonomous Region, China -
About author:Yuan Changshen, Master, Orthopedic Department of the Limbs, The First Affiliated Hospital of Guangxi University of Chinese Medicine, Nanning 530023, Guangxi Zhuang Autonomous Region, China -
Supported by:National Natural Science Foundation of China, Nos. 82060875 (to YCS) and 82160912 (to DK)
CLC Number:
Cite this article
Yuan Changshen, Liao Shuning, Li Zhe, Wu Siping, Chen Lewei, Liu Jinyi, Li Yanhong, Duan Kan. Machine learning combined with bioinformatics to identify and validate key genes for cellular senescence in osteoarthritis[J]. Chinese Journal of Tissue Engineering Research, 2024, 28(20): 3196-3202.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] HUNTER DJ, BIERMA-ZEINSTRA S. Osteoarthritis. Lancet. 2019;393(10182):1745-1759. [2] LÓPEZ-OTÍN C, BLASCO MA, PARTRIDGE L, et al. The hallmarks of aging. Cell. 2013; 153(6):1194-1217. [3] HE S, SHARPLESS NE. Senescence in Health and Disease. Cell. 2017;169(6):1000-1011. [4] GUILAK F, NIMS RJ, DICKS A, et al. Osteoarthritis as a disease of the cartilage pericellular matrix. Matrix Biol. 2018;71-72:40-50. [5] KHOSLA S, FARR JN, TCHKONIA T, et al. The role of cellular senescence in ageing and endocrine disease. Nat Rev Endocrinol. 2020;16(5):263-275. [6] HOU A, CHEN P, TANG H, et al. Cellular senescence in osteoarthritis and anti-aging strategies. Mech Ageing Dev. 2018;175:83-87. [7] GREENER JG, KANDATHIL SM, MOFFAT L, et al. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 2022;23(1):40-55. [8] AVELAR RA, ORTEGA JG, TACUTU R, et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21(1):91. [9] XU Z, KE T, ZHANG Y, et al. Danshensu inhibits the IL-1β-induced inflammatory response in chondrocytes and osteoarthritis possibly via suppressing NF-κB signaling pathway. Mol Med. 2021;27(1):80. [10] ZHU H, YAN X, ZHANG M, et al. miR-21-5p protects IL-1β-induced human chondrocytes from degradation. J Orthop Surg Res. 2019;14(1):118. [11] 魏丽杰.降钙素对IL-1β诱导的大鼠软骨细胞炎性反应的影响[D].唐山:河北联合大学,2014. [12] 中华医学会骨科学分会关节外科学组,中国医师协会骨科医师分会骨关节炎学组,国家老年疾病临床医学研究中心(湘雅医院),等.中国骨关节炎诊疗指南(2021年版)[J].中华骨科杂志,2021,41(18):1291-1314. [13] CORYELL PR, DIEKMAN BO, LOESER RF. Mechanisms and therapeutic implications of cellular senescence in osteoarthritis. Nat Rev Rheumatol. 2021;17(1):47-57. [14] JEON OH, DAVID N, CAMPISI J, et al. Senescent cells and osteoarthritis: a painful connection. J Clin Invest. 2018;128(4):1229-1237. [15] YOON DS, LEE KM, CHOI Y, et al. TLR4 downregulation by the RNA-binding protein PUM1 alleviates cellular aging and osteoarthritis. Cell Death Differ. 2022;29(7):1364-1378. [16] JI ML, JIANG H, LI Z, et al. Sirt6 attenuates chondrocyte senescence and osteoarthritis progression. Nat Commun. 2022;13(1):7658. [17] NEDUNCHEZHIYAN U, VARUGHESE I, SUN AR, et al. Obesity, Inflammation, and Immune System in Osteoarthritis. Front Immunol. 2022;13:907750. [18] WOODELL-MAY JE, SOMMERFELD SD. Role of Inflammation and the Immune System in the Progression of Osteoarthritis. J Orthop Res. 2020;38(2):253-257. [19] GSCHWANDTNER M, DERLER R, MIDWOOD KS. More Than Just Attractive: How CCL2 Influences Myeloid Cell Behavior Beyond Chemotaxis. Front Immunol. 2019;10:2759. [20] GRIESHABER-BOUYER R, KÄMMERER T, ROSSHIRT N, et al. Divergent Mononuclear Cell Participation and Cytokine Release Profiles Define Hip and Knee Osteoarthritis. J Clin Med. 2019;8(10):1631. [21] HAUBRUCK P, PINTO MM, MORADI B, et al. Monocytes, Macrophages, and Their Potential Niches in Synovial Joints - Therapeutic Targets in Post-Traumatic Osteoarthritis? Front Immunol. 2021;12:763702. [22] KLEIN-WIERINGA IR, DE LANGE-BROKAAR BJ, YUSUF E, et al. Inflammatory Cells in Patients with Endstage Knee Osteoarthritis: A Comparison between the Synovium and the Infrapatellar Fat Pad. J Rheumatol. 2016;43(4):771-778. [23] ROSSHIRT N, TRAUTH R, PLATZER H, et al. Proinflammatory T cell polarization is already present in patients with early knee osteoarthritis. Arthritis Res Ther. 2021;23(1):37. [24] NEES TA, ROSSHIRT N, ZHANG JA, et al. T Helper Cell Infiltration in Osteoarthritis-Related Knee Pain and Disability. J Clin Med. 2020;9(8):2423. [25] ZHAO X, ZHAO Y, JIANG Y, et al. Deciphering the endometrial immune landscape of RIF during the window of implantation from cellular senescence by integrated bioinformatics analysis and machine learning. Front Immunol. 2022;13:952708. [26] MARTINEZ E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol. 2002;50(6):925-947. [27] ZHANG M, LU Q, BUDDEN T, et al. NFAT1 protects articular cartilage against osteoarthritic degradation by directly regulating transcription of specific anabolic and catabolic genes. Bone Joint Res. 2019;8(2):90-100. [28] ZHANG Z, ZHANG R. Epigenetics in autoimmune diseases: Pathogenesis and prospects for therapy. Autoimmun Rev. 2015;14(10):854-863. [29] GU XD, WEI L, LI PC, et al. Adenovirus-mediated transduction with Histone Deacetylase 4 ameliorates disease progression in an osteoarthritis rat model. Int Immunopharmacol. 2019;75:105752. [30] BALOUN J, KROPÁČKOVÁ T, HULEJOVÁ H, et al. Chemokine and Cytokine Profiles in Patients with Hand Osteoarthritis. Biomolecules. 2020;11(1):4. [31] NI F, ZHANG Y, PENG X, et al. Correlation between osteoarthritis and monocyte chemotactic protein-1 expression: a meta-analysis. J Orthop Surg Res. 2020;15(1):516. [32] JI T, CHEN M, SUN W, et al. JAK-STAT signaling mediates the senescence of cartilage-derived stem/progenitor cells. J Mol Histol. 2022;53(4):635-643. [33] QIAO Z, TANG J, WU W, et al. Acteoside inhibits inflammatory response via JAK/STAT signaling pathway in osteoarthritic rats. BMC Complement Altern Med. 2019;19(1):264. [34] SUN P, XUE Y. Silence of TANK-binding kinase 1 (TBK1) regulates extracellular matrix degradation of chondrocyte in osteoarthritis by janus kinase (JAK)-signal transducer of activators of transcription (STAT) signaling. Bioengineered. 2022;13(1):1872-1879. [35] LIU W, CHEN Y, ZENG G, et al. INSR mediated by transcription factor KLF4 and DNA methylation ameliorates osteoarthritis progression via inactivation of JAK2/STAT3 signaling pathway. Am J Transl Res. 2020;12(12):7953-7967. [36] SUN K, LUO J, GUO J, et al. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis Cartilage. 2020;28(4):400-409. [37] SUN EJ, WANKELL M, PALAMUTHUSINGAM P, et al. Targeting the PI3K/Akt/mTOR Pathway in Hepatocellular Carcinoma. Biomedicines. 2021;9(11):1639. [38] MATSUZAKI T, ALVAREZ-GARCIA O, MOKUDA S, et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med. 2018;10(428):eaan0746. [39] GUO X, PAN X, WU J, et al. Calycosin prevents IL-1β-induced articular chondrocyte damage in osteoarthritis through regulating the PI3K/AKT/FoxO1 pathway. In Vitro Cell Dev Biol Anim. 2022;58(6):491-502. [40] LI S, LIU J, LIU S, et al. Chitosan oligosaccharides packaged into rat adipose mesenchymal stem cells-derived extracellular vesicles facilitating cartilage injury repair and alleviating osteoarthritis. J Nanobiotechnology. 2021;19(1):343. [41] XU W, WANG X, LIU D, et al. Identification and validation of hub genes and potential drugs involved in osteoarthritis through bioinformatics analysis. Front Genet. 2023;14:1117713. [42] DHANASEKARAN R, DEUTZMANN A, MAHAUAD-FERNANDEZ WD, et al. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. 2022;19(1):23-36. [43] DUFFY MJ, O’GRADY S, TANG M, et al. MYC as a target for cancer treatment. Cancer Treat Rev. 2021;94:102154. [44] VIDOVIĆ T, EWALD CY. Longevity-Promoting Pathways and Transcription Factors Respond to and Control Extracellular Matrix Dynamics During Aging and Disease. Front Aging. 2022;3:935220. [45] LIU J, HUANG X, LIU D, et al. Demethyleneberberine induces cell cycle arrest and cellular senescence of NSCLC cells via c-Myc/HIF-1α pathway. Phytomedicine. 2021;91:153678. [46] BOLESTA E, PFANNENSTIEL LW, DEMELASH A, et al. Inhibition of Mcl-1 promotes senescence in cancer cells: implications for preventing tumor growth and chemotherapy resistance. Mol Cell Biol. 2012;32(10):1879-1892. [47] LU Y, ZHANG C, JIANG S, et al. Anti-Dlx5 Retards the Progression of Osteoarthritis through Inhibiting Chondrocyte Hypertrophy and Apoptosis. Evid Based Complement Alternat Med. 2022;2022:5019920. [48] YATSUGI N, TSUKAZAKI T, OSAKI M, et al. Apoptosis of articular chondrocytes in rheumatoid arthritis and osteoarthritis: correlation of apoptosis with degree of cartilage destruction and expression of apoptosis-related proteins of p53 and c-myc. J Orthop Sci. 2000;5(2):150-156. [49] 陈伟达,石玮,杨继国,等.三七总皂苷经miR-24靶向C-myc对骨关节炎模型大鼠软骨细胞凋亡与增殖的影响[J].浙江中西医结合杂志,2021,31(4):319-323. |
| [1] | Li Yongjie, Fu Shenyu, Xia Yuan, Zhang Dakuan, Liu Hongju. Correlation of knee extensor muscle strength and spatiotemporal gait parameters with peak knee flexion/adduction moment in female patients with knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(9): 1354-1358. |
| [2] | Qi Haodong, Lu Chao, Xu Hanbo, Wang Mengfei, Hao Yangquan. Effect of diabetes mellitus on perioperative blood loss and pain after primary total knee arthroplasty [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(9): 1383-1387. |
| [3] | Du Changling, Shi Hui, Zhang Shoutao, Meng Tao, Liu Dong, Li Jian, Cao Heng, Xu Chuang. Efficacy and safety of different applications of tranexamic acid in high tibial osteotomy [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(9): 1409-1413. |
| [4] | Yu Weijie, Liu Aifeng, Chen Jixin, Guo Tianci, Jia Yizhen, Feng Huichuan, Yang Jialin. Advantages and application strategies of machine learning in diagnosis and treatment of lumbar disc herniation [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(9): 1426-1435. |
| [5] | Huang Xiarong, Hu Lizhi, Sun Guanghua, Peng Xinke, Liao Ying, Liao Yuan, Liu Jing, Yin Linwei, Zhong Peirui, Peng Ting, Zhou Jun, Qu Mengjian. Effect of electroacupuncture on the expression of P53 and P21 in articular cartilage and subchondral bone of aged rats with knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(8): 1174-1179. |
| [6] | Zhao Garida, Ren Yizhong, Han Changxu, Kong Lingyue, Jia Yanbo. Mechanism of Mongolian Medicine Erden-uril on osteoarthritis in rats [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(8): 1193-1199. |
| [7] | Li Rui, Zhang Guihong, Wang Tao, Fan Ping. Effect of ginseng polysaccharide on the expression of prostaglandin E2/6-keto-prostaglandin 1alpha in traumatic osteoarthritis model rats [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(8): 1235-1240. |
| [8] | Zhang Kefan, Shi Hui. Research status and application prospect of cytokine therapy for osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(6): 961-967. |
| [9] | Zhang Zeyi, Yang Yimin, Li Wenyan, Zhang Meizhen. Effect of foot progression angle on lower extremity kinetics of knee osteoarthritis patients of different ages: a systematic review and meta-analysis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(6): 968-975. |
| [10] | Shen Feiyan, Yao Jixiang, Su Shanshan, Zhao Zhongmin, Tang Weidong. Knockdown of circRNA WD repeat containing protein 1 inhibits proliferation and induces apoptosis of chondrocytes in knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(4): 499-504. |
| [11] | Maisituremu·Heilili, Zhang Wanxia, Nijiati·Nuermuhanmode, Maimaitituxun·Tuerdi. Effect of intraarticular injection of different concentrations of ozone on condylar histology of rats with early temporomandibular joint osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(4): 505-509. |
| [12] | Qiao Hujun, Wang Guoxiang. Evaluation of rat osteoarthritis chondrocyte models induced by interleukin-1beta [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(4): 516-521. |
| [13] | Bu Xianzhong, Bu Baoxian, Xu Wei, Zhang Chi, Zhang Yisheng, Zhong Yuanming, Li Zhifei, Tang Fubo, Mai Wei, Zhou Jinyan. Analysis of serum differential proteomics in patients with acute cervical spondylotic radiculopathy [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(4): 535-541. |
| [14] | Liu Yuhan, Fan Yujiang, Wang Qiguang. Comparison of protocols for constructing animal models of early traumatic knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(4): 542-549. |
| [15] | Zhang Yaru, Chen Yanjun, Zhang Xiaodong, Chen Shenghua, Huang Wenhua. Effect of ferroptosis mediated by glutathione peroxidase 4 in the occurrence and progression of synovitis in knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(4): 550-555. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||