Chinese Journal of Tissue Engineering Research ›› 2022, Vol. 26 ›› Issue (19): 3011-3017.doi: 10.12307/2022.379
Previous Articles Next Articles
Heterogeneity of chondrocytes derived from human ribs based on single-cell transcriptome sequencing
Shi Hang, Li Jia, Liu Xia, Jiang Haiyue
- Research Center of Plastic Surgery Hospital, Chinese Academy of Medical Science & Peking Union Medical College, Beijing 100144, China
-
Received:2021-03-23Revised:2021-04-28Accepted:2021-08-04Online:2022-07-08Published:2021-12-28 -
Contact:Liu Xia, MD, Researcher, Research Center of Plastic Surgery Hospital, Chinese Academy of Medical Science & Peking Union Medical College, Beijing 100144, China -
About author:Shi Hang, Master, Research Center of Plastic Surgery Hospital, Chinese Academy of Medical Science & Peking Union Medical College, Beijing 100144, China -
Supported by:General Program of the National Natural Science Foundation of China, No. 81871575 (to LX); Major Collaborative Innovation Project of Medical and Health Technology Innovation Project of Chinese Academy of Medical Sciences, No. 2017-I2M-1-007 (to JHY)
CLC Number:
Cite this article
Shi Hang, Li Jia, Liu Xia, Jiang Haiyue. Heterogeneity of chondrocytes derived from human ribs based on single-cell transcriptome sequencing[J]. Chinese Journal of Tissue Engineering Research, 2022, 26(19): 3011-3017.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
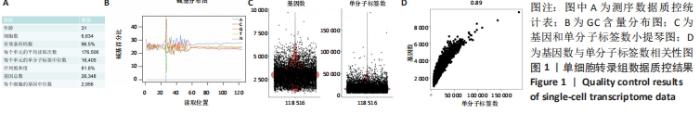
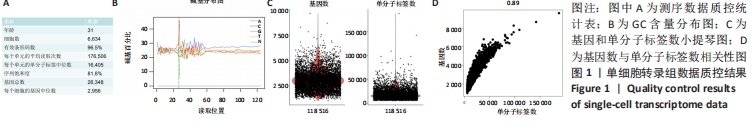
2.1 10X Genomics样品处理和cDNA文库制备 人肋软骨组织制备成单细胞悬液,使用Countess?Ⅱ Automated Cell Counter进行细胞计数和细胞活率测定,细胞活率为85%,将细胞浓度调整为1×109 L-1。利用Illumina HiSeq测序平台进行测序和初级分析,进行数据可视化,在单细胞水平上生成了肋软骨组织中基因表达的深层转录图,获得6 634个细胞进行分析,检测到单个细胞的单分子标签和基因中位数分别为16 405和2 956。81.8%的测序饱和率表明对可用转录本进行了全面取样。以上数据见图1A。 对数据分析之前进行了严格的数据质控,首先检测了碱基含量的分布,见图1B,其用于检测有无AT、GC分离现象。理论上普通文库的G和C碱基及A和T碱基含量每个测序循环上应分别相等,且整个测序过程稳定不变,呈水平线。由于随机引物扩增偏差等原因,常常会导致在测序得到的每个read前几个碱基有较大的波动,这种波动属于正常情况。由图1B中GC含量分布看出,未检测到AT、GC分离现象。在文库构建过程中可能会掺入死细胞,或者多个细胞被捕获在同一个液滴中,产生潜在的多重细胞,因此利用基因数和单分子标签数进行质控,见图1C,基因数大于5 000、大部分细胞的单分子标签数大于10 000,是一组高质量的测序数据。随着测序数据量的增加,单细胞检测到的基因数量和单分子标签的数量也随之增加,两者之间呈明显正相关,见图1D。细胞的单分子标签数与基因数的相关性可以确定是否有大量死亡或濒死细胞的线粒体污染,此次数据得分为0.89,符合要求,可以进行后续分析。"
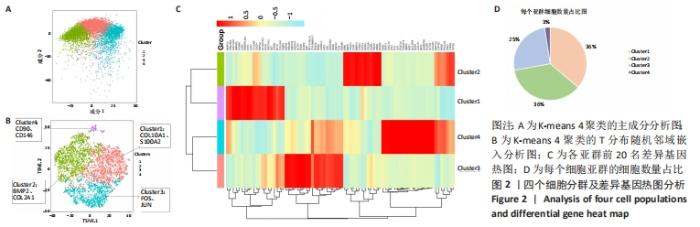
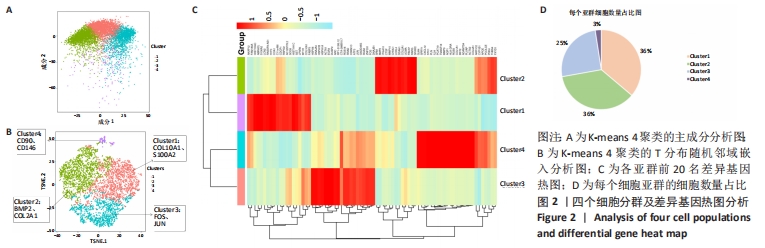
2.2 根据单细胞转录组测序数据进行细胞分群 为了深入剖析肋软骨细胞的异质性,首先对表达数据单分子标签归一化,然后主成分分析降维,见图2A,选取K-means聚类算法进行细胞分群,同时利用T分布随机邻域嵌入进行可视化的聚类分析,获得了4个聚类(Cluster)的细胞群,不同类的细胞亚群标记为不同的颜色,见图2B。每个亚群都有特异的标记基因,图中显示了各亚群中前20名的差异基因,见图2C。结合软骨细胞特征性基因在各群中的表达情况,发现Cluster1是以COL10A1、S100A2为标记基因的细胞群,这群细胞在该样本中占比36%;Cluster2是以BMP2、COL2A1基因为标记基因的细胞群,细胞数占总样本的36%;Cluster3是以FOS、JUN基因为标记基因的细胞群,细胞数占总样本的25%;Cluster4是以CD90、CD146基因为标记基因的细胞群,细胞数占总样本的数量最少,仅为3%,见图2D。"
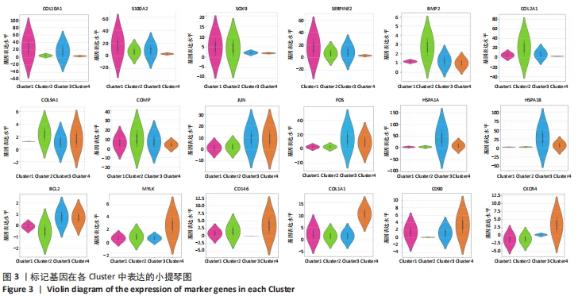
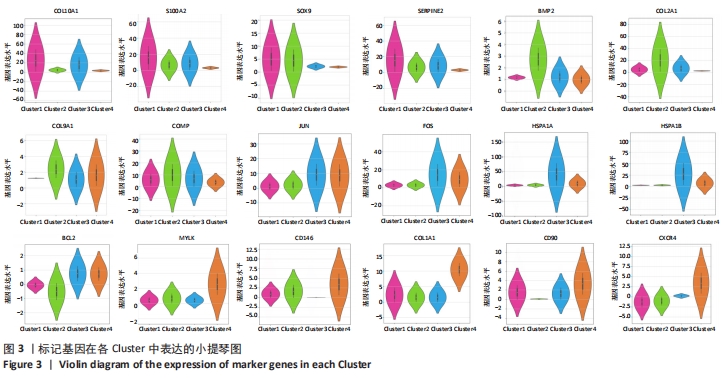
为了进一步区分每个聚类,基于基因平均表达量和亚群之间的差异表达基因进行协方差统计分析,研究发现标记基因均具有较高的特异性,可以识别细胞类型,见图3。Cluster1中肥大软骨基因COL10A1、钙调蛋白S100A2、抑制基质降解蛋白SERPINE2、软骨标记基因SOX9表达比较高,成软骨基因COL2A1、COL9A1、增殖调控基因FOS、JUN、BCL2,热调蛋白基因HSPA1A表达相对较低,因此这群细胞高表达肥大软骨细胞标记基因,将Cluster1注释为肥大软骨细胞群。Cluster2中成软骨基因COL2A1、COL9A1、SOX9、BMP2,钙调蛋白S100A2的基因表达相对较高,增殖性基因FOS、JUN,热调蛋白HSPA1A,干细胞基因CD90、CD146的表达均比较低,因此Cluster2是一群向软骨分化完全,但增殖能力不强的软骨细胞群。Cluster3中增殖性基因FOS、JUN,抑制凋亡的基因BCL2,调控细胞生长的基因HSPA1A、HAPA1B特异性高表达,软骨分化基因SOX9、COL2A1,干细胞基因CD146在Cluster3的表达相对较少,因此Cluster3被定义为一类增殖性细胞群。Cluster4中肥大软骨基因COL10A1,成软骨基因SOX9、COL2A1表达均较少,但调控细胞发育相关的基因MYLK、干细胞标记基因CD146、CD90等在Cluster4中特异性表达,将Cluster4注释为干细胞群。"
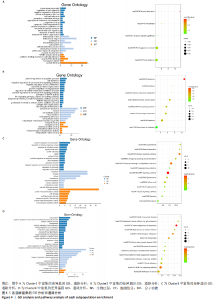
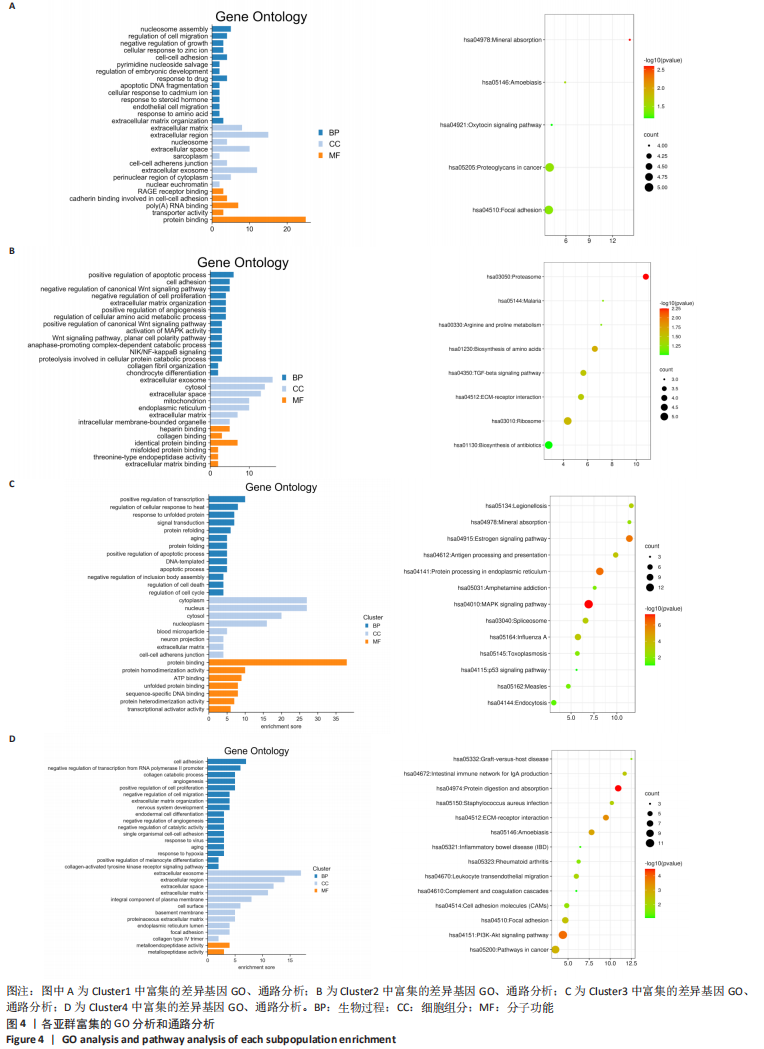
2.3 细胞亚群标记基因参与的生物过程 为了进一步研究肋软骨组织中各细胞亚群相关的功能状态和潜在的调控因子,对每个细胞群中富集的差异表达基因进行GO和KEGG分析,见图4,结果显示:在被注释为肥大软骨细胞群的Cluster 1中,生物学功能方面主要富集细胞间的黏附和对细胞迁移的调控;在细胞组分方面主要富集在细胞外基质、细胞外泌体之中;在分子功能方面主要富集蛋白质结合和细胞黏附等功能。在被注释为软骨细胞群的Cluster 2中,生物学功能方面主要富集软骨细胞分化、Wnt信号通路的调节、细胞增殖和凋亡的调节;在细胞组分方面主要富集在细胞外泌体,细胞外基质中;在分子功能方面主要富集胶原结合、蛋白质结合、细胞外基质结合等功能。在被注释为增殖性软骨细胞群的Cluster 3中,生物学功能主要富集信号传导、细胞周期调控等方面;细胞组分方面主要富集在细胞质、细胞核、细胞外基质等部位;在分子功能方面主要富集蛋白结合、ATP结合等方面。在被注释为干细胞的Cluster 4中,生物学功能主要富集在细胞增殖、细胞分化、细胞黏附、胶原分解代谢等方面;细胞组分主要在细胞外基质、细胞外泌体、细胞膜等部位;分子功能主要富集在激活金属酶等方面。综上所述,在细胞组分方面4个亚群都主要富集在细胞外基质,在分子功能方面都主要与蛋白质结合有关。"
| [1] VILA PM, JEANPIERRE LM, RIZZI CJ, et al. Comparison of Autologous vs Homologous Costal Cartilage Grafts in Dorsal Augmentation Rhinoplasty: A Systematic Review and Meta-analysis. JAMA Otolaryngol Head Neck Surg. 2020;146(4):347-354. [2] CHOI SW, HONG KY, MINN KW, et al. Chondrogenesis of Adipose-Derived Stem Cells on Irradiated Cartilage. Plast Reconstr Surg. 2020; 145(2):409-418. [3] JUNG SN, RHIE JW, KWON H, et al. In vivo cartilage formation using chondrogenic-differentiated human adipose-derived mesenchymal stem cells mixed with fibrin glue. J Craniofac Surg. 2010;21(2):468-472. [4] ZHAO M, CHEN Z, LIU K, et al. Repair of articular cartilage defects in rabbits through tissue-engineered cartilage constructed with chitosan hydrogel and chondrocytes. J Zhejiang Univ Sci B. 2015;16(11):914-923. [5] SAN-MARINA S, SHARMA A, VOSS SG, et al. Assessment of Scaffolding Properties for Chondrogenic Differentiation of Adipose-Derived Mesenchymal Stem Cells in Nasal Reconstruction. JAMA Facial Plast Surg. 2017;19(2):108-114. [6] DION J, COSTEDOAT-CHALUMEAU N, SÈNE D, et al. Relapsing Polychondritis Can Be Characterized by Three Different Clinical Phenotypes: Analysis of a Recent Series of 142 Patients. Arthritis Rheumatol. 2016;68(12):2992-3001. [7] ENOMURA M, MURATA S, TERADO Y, et al. Development of a Method for Scaffold-Free Elastic Cartilage Creation. Int J Mol Sci. 2020;21(22): 8496. [8] 靳小兵.自体组织工程生长板修复兔长骨生长板损伤的实验研究[D].西安:第四军医大学,2004. [9] 姜疆.在体构建组织工程软骨的实验研究[D].西安:第四军医大学,2012. [10] CHEN R, WU X, JIANG L, et al. Single-Cell RNA-Seq Reveals Hypothalamic Cell Diversity. Cell Rep. 2017;18(13):3227-3241. [11] HWANG B, LEE JH, BANG D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50(8):1-14. [12] GULATI GS, SIKANDAR SS, WESCHE DJ, et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science. 2020;367 (6476):405-411. [13] MAHMOUDI S, MANCINI E, XU L, et al. Heterogeneity in old fibroblasts is linked to variability in reprogramming and wound healing. Nature. 2019;574(7779):553-558. [14] QI F, QIAN S, ZHANG S, et al. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526(1):135-140. [15] JI Q, ZHENG Y, ZHANG G, et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis. 2019;78(1):100-110. [16] SUN H, WEN X, LI H, et al. Single-cell RNA-seq analysis identifies meniscus progenitors and reveals the progression of meniscus degeneration. Ann Rheum Dis. 2020;79(3):408-417. [17] 胡坤,陈竹,罗栩伟,等.组织工程软骨的研究新进展[J].西部医学, 2020,32(6):927-932. [18] FU S, KUWAHARA M, UCHIDA Y, et al. Circadian production of melatonin in cartilage modifies rhythmic gene expression. J Endocrinol. 2019:JOE-19-0022.R2. [19] MEI R, LOU P, YOU G, et al. 17β-Estradiol Induces Mitophagy Upregulation to Protect Chondrocytes via the SIRT1-Mediated AMPK/mTOR Signaling Pathway. Front Endocrinol (Lausanne). 2021;11:615250. [20] 杨美蓉.先天性小耳畸形伴发胸廓畸形的临床研究及初步遗传学分析[D].北京:协和医学院,2018. [21] PRETEMER Y, KAWAI S, NAGATA S, et al. Differentiation of Hypertrophic Chondrocytes from Human iPSCs for the In Vitro Modeling of Chondrodysplasias. Stem Cell Reports. 2021;16(3):610-625. [22] 薛松,姜亚飞,桑伟林,等.肥大软骨细胞在骨关节炎发病中的作用[J].中国矫形外科杂志,2020,28(6):522-526. [23] 陈海涛,倪曲波,陈廖斌.关节软骨的发育及调控[J].武汉大学学报(医学版),2021,42(2):333-337. [24] SANTORO A, CONDE J, SCOTECE M, et al. SERPINE2 Inhibits IL-1α-Induced MMP-13 Expression in Human Chondrocytes: Involvement of ERK/NF-κB/AP-1 Pathways. PLoS One. 2015;10(8):e0135979. [25] DANG QT, HUYNH TD, INCHINGOLO F, et al. Human Chondrocytes from Human Adipose Tissue-Derived Mesenchymal Stem Cells Seeded on a Dermal-Derived Collagen Matrix Sheet: Our Preliminary Results for a Ready to Go Biotechnological Cartilage Graft in Clinical Practice. Stem Cells Int. 2021;2021:6664697. [26] ZHAO C, WANG E. Heat shock protein 90 suppresses tumor necrosis factor alpha induced apoptosis by preventing the cleavage of Bid in NIH3T3 fibroblasts. Cell Signal. 2004;16(3):313-321. [27] 杜俊杰,罗卓荆,胡蕴玉,等.rhBMP-2在体内诱导成骨中I,II型胶原及碱性磷酸酶的表达[J].第四军医大学学报,2001,22(11):981-983. [28] 张艳,柴岗,刘伟,等.人肋软骨细胞体外培养中生长代谢及功能的变化[J].中华整形外科杂志,2004,20(5):372-376. [29] IANNONE F, CORRADO A, GRATTAGLIANO V, et al. Phenotyping of chondrocytes from human osteoarthritic cartilage: chondrocyte expression of beta integrins and correlation with anatomic injury. Reumatismo. 2001;53(2):122-130. [30] KURKOV A, GULLER A, FAYZULLIN A, et al. Amianthoid transformation of costal cartilage matrix in children with pectus excavatum and pectus carinatum. PLoS One. 2021;16(1):e0245159. [31] KATO Y, GOSPODAROWICZ D. Effect of exogenous extracellular matrices on proteoglycan synthesis by cultured rabbit costal chondrocytes. J Cell Biol. 1985;100(2):486-495. [32] SATO K, MOY OJ, PEIMER CA, et al. An experimental study on costal osteochondral graft. Osteoarthritis Cartilage. 2012;20(2):172-183. [33] 赵琴琴,蔡震,游晓波,等.不同性别不同层次肋软骨的生物力学性能研究[J].中华整形外科杂志,2020,36(9):1042-1046. [34] FORMAN JL, KENT RW. The effect of calcification on the structural mechanics of the costal cartilage. Comput Methods Biomech Biomed Engin. 2014;17(2):94-107. [35] KIM A, MOON J, LIM SY, et al. The Interchondral Joints of Thorax in Microtia Surgery: Classification and Fabrication Strategies. Ann Plast Surg. 2021 Feb 1. doi: 10.1097/SAP.0000000000002582. [36] 周志文,鞠黎,楼跃.不同年龄软骨组织多肽的定量分析[J].中国组织工程研究,2018,22(8):1178-1183. [37] 吴迎,王先成,熊祥,等.70例女性第6-8肋软骨组织量及钙化特点分析[J].中华整形外科杂志,2019,35(8):764-771. [38] GUO F, YU X, CHEN W, et al. Preliminary Analysis on Characteristics of Rib Cartilage Calcification in Patients With Congenital Microtia. J Craniofac Surg. 2019;30(1):e28-e32. |
| [1] | Guan Hong, Zhang Hongbo, Shao Yan, Guo Dong, Zhang Haiyan, Cai Daozhang. PDZ domain containing 1 deficiency promotes chondrocyte senescence in osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(2): 182-189. |
| [2] | Ma Dujun, Peng Liping, Zhou Ziqiong, Zhao Jing, Zhu Houjun, Jiang Shunwan, Zhong Jing, She Ruihao. Effect of RAB39B gene based on CRISPR/Cas9 technology on cartilage differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(19): 2978-2984. |
| [3] | Yang Shuang, Yan Jinglong. Effects of SOX9 on chondrocyte differentiation [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(14): 2279-2284. |
| [4] | Huang Chang, Ying Peixi, Yi Guoguo, Fu Sheng, Wang Yan, Guo Xi, Zhang Yuxi, Fu Min. Application of single-cell RNA sequencing in diagnosis and treatment of corneal and retinal diseases [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(13): 2106-2113. |
| [5] | Wang Hailong, Li Long, Wang Jianyuan, Yan Bin, Xier Aili, Chen Hongtao, Yilihamu · Tuoheti. High-throughput sequencing-based osteoporotic circRNA-miRNA-mRNA network construction and Has_ circ_0076906 functional verification [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(11): 1705-1713. |
| [6] | Feng Zhiguo, Sun Haibiao, Han Xiaoqiang. Regulation of proliferation, differentiation and apoptosis of bone-related cells by long-stranded non-coding RNA [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(1): 112-118. |
| [7] | Liu Xiaogang, Li Tian, Zhang Duo. Effect and mechanism of the effective components of Chinese medicine on promoting the differentiation of bone marrow mesenchymal stem cells into chondrocytes [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(1): 119-124. |
| [8] | Xie Chongxin, Zhang Lei. Comparison of knee degeneration after anterior cruciate ligament reconstruction with or without remnant preservation [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(5): 735-740. |
| [9] | Ma Zetao, Zeng Hui, Wang Deli, Weng Jian, Feng Song. MicroRNA-138-5p regulates chondrocyte proliferation and autophagy [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(5): 674-678. |
| [10] | Zhang Yiyun, Wang Ziqiang, Ren Ying, Du Fuchong, Du Bo, Li Xuemin. Effect of modified citrus pectin on chondrocytes [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(34): 5465-5472. |
| [11] | Liu Guanjuan, Song Na, An Zheqing, Sun Jing, Liao Jian. Metagenomics and peri-implantitis [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(34): 5511-5516. |
| [12] | Liu Xiaogang, Li Tian, Zhang Duo. Research hotspot of seed cell selection in cartilage tissue engineering [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(31): 5059-5064. |
| [13] | Chen Pu, Ruan Anmin, Zhou Jun, Zhang Xiaozhe , Ma Yufeng, Peng Haoxuan, Yang Tongjie, Wang Qingpu. Effect of total glucosides of paeony on inflammation and degeneration of chondrocytes in osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(29): 4614-4618. |
| [14] | Wang Xianggang, Wan Qian, Liu He, Li Ronghang, Zhang Yan, Li Zuhao, Wang Jincheng. Role and characteristics of tissue-engineered cartilage in the treatment of growth plate injuries [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(28): 4539-4545. |
| [15] | Zhang Chuanhui, Li Jianjun, Yang Jun. Effect of dynamic compression combined with insulin-like growth factor-1 gene transfection on chondrogenic differentiation of rabbit adipose-derived mesenchymal stem cells in chitosan/gelatin scaffolds [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(28): 4485-4491. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||