Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (5): 1147-1155.doi: 10.12307/2026.022
Previous Articles Next Articles
Autophagy regulates early embryonic development in mice via affecting H3K4me3 modification
Hu Jing1, Zhu Ling2, Xie Juan2, Kong Deying1, Liu Doudou2
- 1Department of Physiology, School of Basic Medical Sciences, Zunyi Medical University, Zunyi 563099, Guizhou Province, China; 2Chongqing Key Laboratory of Human Embryo Engineering, Chongqing Health Center for Women and Children, Chongqing 400013, China
-
Received:2024-11-02Accepted:2024-12-28Online:2026-02-18Published:2025-06-24 -
Contact:Liu Doudou, PhD, Chongqing Key Laboratory of Human Embryo Engineering, Chongqing Health Center for Women and Children, Chongqing 400013, China Co-corresponding author: Kong Deying, PhD, Associate professor, Department of Physiology, School of Basic Medical Sciences, Zunyi Medical University, Zunyi 563099, Guizhou Province, China -
About author:Hu Jing, Master candidate, Department of Physiology, School of Basic Medical Sciences, Zunyi Medical University, Zunyi 563099, Guizhou Province, China -
Supported by:a grant from Chongqing Science and Technology Bureau, No. CSTB2022NSCQ-MSX0875 (to XJ [project participant]); National Natural Science Foundation of China, No. 31760339 (to KDY [project participant]); Guizhou Science and Technology Program Project, No. Qiankeheji ZK(2022)General 594 (to KDY [project participant])
CLC Number:
Cite this article
Hu Jing, Zhu Ling, Xie Juan, Kong Deying, Liu Doudou. Autophagy regulates early embryonic development in mice via affecting H3K4me3 modification[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1147-1155.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
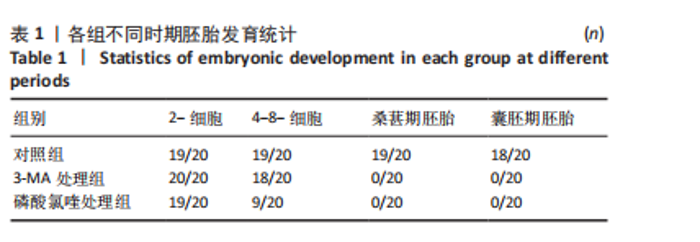
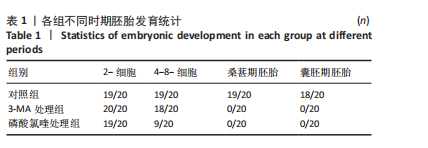
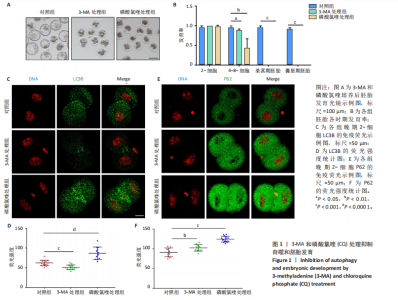
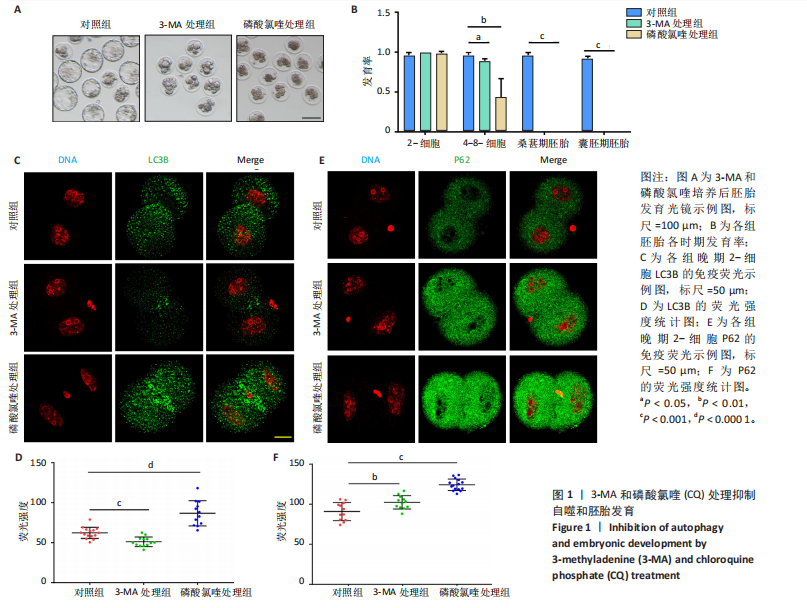
2.1 自噬抑制影响早期胚胎发育 为了评估自噬抑制后对胚胎发育的影响,将小鼠受精卵分别放入含3-MA和磷酸氯喹的培养液中培养。结果显示,3-MA处理组(n=20)所有胚胎发育都没有超过4-8-细胞阶段(P < 0.001),而磷酸氯喹处理组(n=20)的胚胎50%停滞在2-细胞阶段(P < 0.01),45%在4-8-细胞阶段阻滞(P < 0.001),见图1A,B和表1。对晚期2-细胞自噬相关蛋白的免疫荧光检测结果显示,3-MA处理组(n=13)的LC3B水平相对对照组(n=15)显著下降(P < 0.001),磷酸氯喹处理组(n=12)的LC3B水平明显升高(P < 0.000 1),见图1C,D。此外,P62蛋白水平在3-MA处理组(n=12,P < 0.01)和磷酸氯喹处理组(n=15,P < 0.001)中均明显升高,见图1E,F。以上结果表明自噬抑制会造成胚胎发育阻滞。"
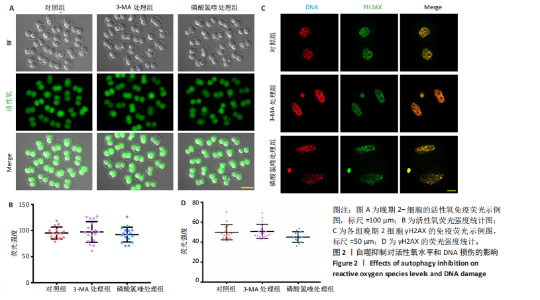
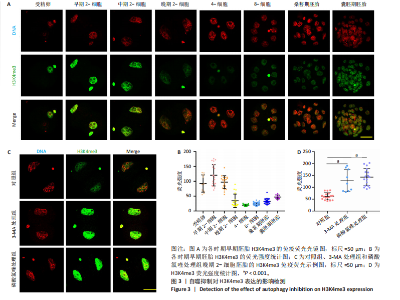
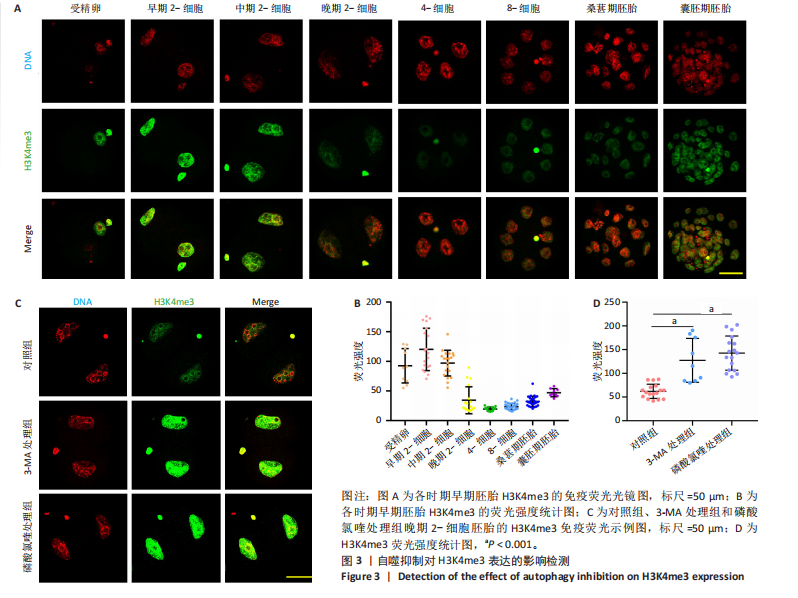
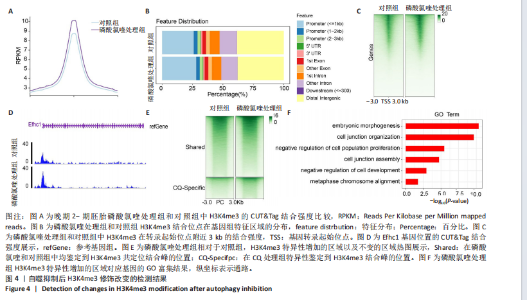
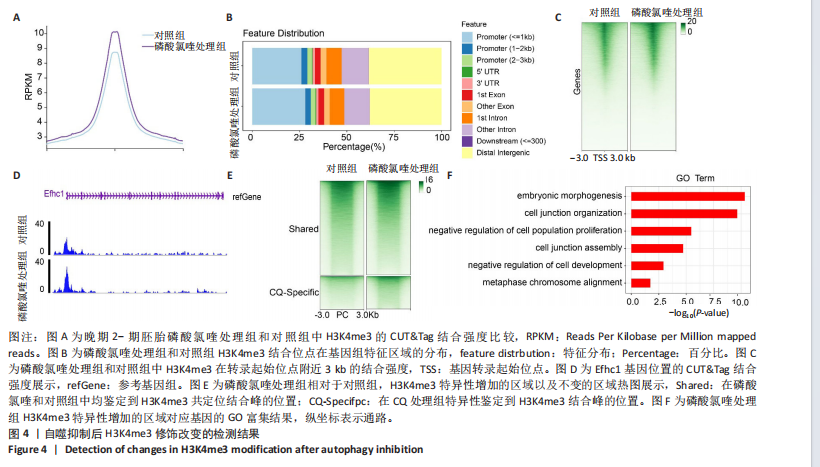
2.2 自噬抑制不诱导胚胎的活性氧和DNA损伤 3-MA和磷酸氯喹处理后检测小鼠晚期2-细胞胚胎的活性氧和DNA损伤标记物γH2AX,结果显示,3-MA处理组(n=18)和磷酸氯喹处理组(n=17)的活性氧水平与对照组(n=15)无明显差异(P > 0.05),见图2A,B。γH2AX的免疫染色结果表明,3-MA处理组(n=19)与对照组(n=16)相比没有显著差异(P > 0.05),磷酸氯喹处理组(n=13)相对于对照组(n=16)没有显著差异(P > 0.05),见图2C,D。综合以上结果表明自噬抑制对胚胎发育的影响不是由活性氧和DNA损伤引 起的。 2.3 自噬抑制导致胚胎H3K4me3修饰水平增强 2.3.1 各阶段胚胎的H3K4me3水平 为了明确小鼠早期胚胎发育过程中H3K4me3修饰的动态变化,通过免疫荧光检测了小鼠胚胎从受精卵到囊胚阶段的H3K4me3水平。结果显示,H3K4me3修饰水平从受精卵到早期2-细胞阶段增强,从中期2-细胞到4-细胞阶段逐渐减弱并降到最低;从4-细胞一直到囊胚阶段又逐渐升高,但这一阶段H3K4me3的总体表达量相对于合子到中期2-细胞阶段较低,见图3A,B。 2.3.2 自噬抑制后H3K4me3的表达 为了明确自噬抑制后对H3K4me3修饰是否产生影响,用3-MA和磷酸氯喹处理胚胎后通过免疫荧光检测了晚期2-细胞胚胎的H3K4me3的表达量。结果显示,3-MA(n=9)和磷酸氯喹处理组(n=17)的H3K4me3水平相较于对照组(n=17)均显著升高(P < 0.001),见图3C,D。 以上结果表明,H3K4me3在胚胎发育过程中经历了修饰逐渐减少然后又逐渐增多的动态变化,在晚期2-细胞时期,胚胎的H3K4me3修饰水平降到了较低水平,自噬抑制剂导致该时期胚胎的H3K4me3修饰水平升高,这可能是自噬抑制造成胚胎发育阻滞的原因之一。 2.4 自噬抑制导致胚胎的H3K4me3修饰模式和强度改变 为进一步确定自噬抑制对H3K4me3修饰的影响,进行了CUT&Tag检测分析。结果显示,相较于对照组,磷酸氯喹处理组H3K4me3整体的修饰水平增加(图4A)。通过分类H3K4me3修饰区域,结果显示磷酸氯喹处理后,H3K4me3在近端启动子区域内富集明显增加(图4B),且热图分析和IGV示例图显示,H3K4me3在转录起始位点(TSS)附近修饰增强(图4C,D)。对H3K4me3修饰基因进行分类分析,结果显示磷酸氯喹处理后,H3K4me3修饰水平特异性增加的基因数量显著增多(图4E),对修饰水平特异性增加的基因进行GO功能富集分析显示,这些基因主要参与细胞形态发生、细胞连接和细胞增殖的调控(图4F)。以上结果说明自噬通过改变H3K4me3的修饰模式,影响与细胞形态、细胞连接和细胞增殖等关键生物学过程,从而调控胚胎的正常发育。"
| [1] FENG Y, CHEN Y, WU X, et al. Interplay of energy metabolism and autophagy. Autophagy. 2024;20(1):4-14. [2] BOYA P, CODOGNO P, RODRIGUEZ-MUELA N. Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development. 2018;145(4):dev146506. [3] TABIBZADEH S. Role of autophagy in aging: The good, the bad, and the ugly. Aging Cell. 2023;22(1):e13753. [4] GÓMEZ-VIRGILIO L, SILVA-LUCERO M-DC, FLORES-MORELOS DS, et al. Autophagy: A Key Regulator of Homeostasis and Disease: An Overview of Molecular Mechanisms and Modulators. Cells. 2022;11(15):2262. [5] ALLEN EA, BAEHRECKE EH. Autophagy in animal development. Cell Death Differ. 2020;27(3): 903-918. [6] SONG S, GUO Q, ZHU Y, et al. Exploring the role of autophagy during early human embryonic development through single-cell transcriptome and methylome analyses. Sci China Life Sci. 2022;65(5):940-952. [7] TSUKAMOTO S, KUMA A, MURAKAMI M, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008; 321(5885):117-120. [8] TSUKAMOTO S, KUMA A, MIZUSHIMA N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008;4(8):1076-1078. [9] KOMATSU M, WAGURI S, UENO T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425-434. [10] YUE Z, JIN S, YANG C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077-15082. [11] HONDA BM, DIXON GH, CANDIDO EP. Sites of in vivo histone methylation in developing trout testis. J Biol Chem. 1975;250(22): 8681-8685. [12] ALBERT TK, KERL K. A histone tale that enCOMPASSes pausing: new insights into the functional repertoire of H3K4me3. Signal Transduct Target Ther. 2023;8(1):270. [13] YU H, LESCH B J. Functional Roles of H3K4 Methylation in Transcriptional Regulation. Mol Cell Biol. 2024;44(11):505-515. [14] BARSOUM M, STENZEL AT, BOCHYŃSKA A, et al. Loss of the Ash2l subunit of histone H3K4 methyltransferase complexes reduces chromatin accessibility at promoters. Sci Rep. 2022;12(1):21506. [15] DAHL J A, JUNG I, AANES H, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016; 537(7621):548-552. [16] ZHANG B, ZHENG H, HUANG B, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537(7621):553-557. [17] CAO X, MA T, FAN R, et al. Broad H3K4me3 Domain Is Associated with Spatial Coherence during Mammalian Embryonic Development. bioRxiv [Preprint]. 2023;11:570452. [18] ZHOU C, HALSTEAD MM, BONNET-GARNIER A, et al. Resetting H3K4me3, H3K27ac, H3K9me3 and H3K27me3 during the maternal-to-zygotic transition and blastocyst lineage specification in bovine embryos. bioRxiv. 2022;7:486777. [19] LIU X, WANG C, LIU W, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558-562. [20] DANG Y, LUO L, SHI Y, et al.KDM5-mediated redistribution of H3K4me3 is required for oocyte-to-embryonic transition in cattle†. Biol Reprod. 2022;106(6):1059-1071. [21] ARTAL-MARTINEZ DE NARVAJAS A, GOMEZ TS, ZHANG JS, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33(20):3983-3993. [22] SHIN HJ, KIM H, OH S, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534(7608): 553-557. [23] FÜLLGRABE J, LYNCH-DAY MA, HELDRING N, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500(7463):468-471. [24] TSUKAMOTO S, HARA T, YAMAMOTO A, et al. Fluorescence-based visualization of autophagic activity predicts mouse embryo viability. Sci Rep. 2014;4:4533. [25] SHEN X, ZHANG N, WANG Z, et al. Induction of autophagy improves embryo viability in cloned mouse embryos. Sci Rep. 2015;5:17829. [26] ZHANG L, ZHANG Y, HAN Z, et al. Transcriptome Analyses Reveal Effects of Vitamin C-Treated Donor Cells on Cloned Bovine Embryo Development. Int J Mol Sci. 2019;20(11):2628. [27] XU J, SHI P, ZHAO X, et al. Cell synchronization by Rapamycin improves the developmental competence of buffalos (Bubalus bubalis) somatic cell nuclear transfer embryos. Reprod Domest Anim. 2021;56(2):313-323. [28] DELUAO JC, WINSTANLEY Y, ROBKER RL, et al. OXIDATIVE STRESS AND REPRODUCTIVE FUNCTION: Reactive oxygen species in the mammalian pre-implantation embryo. Reproduction. 2022;164(6):F95-F108. [29] LEITE RF, ANNES K, ISPADA J, et al. Oxidative Stress Alters the Profile of Transcription Factors Related to Early Development on In Vitro Produced Embryos. Oxid Med Cell Longev. 2017;2017:1502489. [30] JAMIL M, DEBBARH H, ABOULMAOUAHIB S, et al. Reactive oxygen species in reproduction: harmful, essential or both?. Zygote. 2020; 28(4):255-269. [31] KEANE JA, EALY AD. An Overview of Reactive Oxygen Species Damage Occurring during In Vitro Bovine Oocyte and Embryo Development and the Efficacy of Antioxidant Use to Limit These Adverse Effects. Animals (Basel). 2024;14(2):330. [32] XU Q, XIE W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 2018;28(3):237-253. [33] WILKINSON AL, ZORZAN I, RUGG-GUNN PJ. Epigenetic regulation of early human embryo development. Cell Stem Cell. 2023;30(12): 1569-1584. [34] 张立苹, 林郑云, 华再东, 等. 组蛋白H3K4me3甲基化修饰与哺乳动物早期胚胎发育[J]. 湖北畜牧兽医,2019,40(7):16-17+19. [35] BU G, ZHU W, LIU X, et al. Coordination of zygotic genome activation entry and exit by H3K4me3 and H3K27me3 in porcine early embryos. Genome Res. 2022;32(8):1487-1501. [36] LI C, ZHANG Y, SHEN J, et al. Cfp1 Controls Cardiomyocyte Maturation by Modifying Histone H3K4me3 of Structural, Metabolic, and Contractile Related Genes. Adv Sci (Weinh). 2024;11(11):e2305992. [37] SU WC, MAO XM, LI SY, et al. DPY30 Promotes Proliferation and Cell Cycle Progression of Colorectal Cancer Cells via Mediating H3K4 Trimethylation. Int J Med Sci. 2023;20(7):901-917. |
| [1] | Liu Kexin, , Hao Kaimin, Zhuang Wenyue, , Li Zhengyi. Autophagy-related gene expression in pulmonary fibrosis models: bioinformatic analysis and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1129-1138. |
| [2] | Yin Yongcheng, Zhao Xiangrui, Yang Zhijie, Li Zheng, Li Fang, Ning Bin. Effect and mechanism of peroxiredoxin 1 in microglial inflammation after spinal cord injury [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1106-1113. |
| [3] | Chen Qiheng, Weng Tujun, Peng Jiang. Effect of dimethylglyoxal glycine on osteogenic, adipogenesis differentiation, and mitophagy of human bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(1): 50-57. |
| [4] | Li Huayuan, Li Chun, Liu Junwei, Wang Ting, Li Long, Wu Yongli. Effect of warm acupuncture on PINK1/Parkin pathway in the skeletal muscle of rats with chronic fatigue syndrome [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1618-1625. |
| [5] | Zhou Panpan, Cui Yinglin, Zhang Wentao, Wang Shurui, Chen Jiahui, Yang Tong . Role of cellular autophagy in cerebral ischemic injury and the regulatory mechanism of traditional Chinese medicine [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1650-1658. |
| [6] | Zhu Hanmin, Wang Song, Xiao Wenlin, Zhang Wenjing, Zhou Xi, He Ye, Li Wei, . Mitophagy regulates bone metabolism [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1676-1683. |
| [7] | Chi Wenxin, Zhang Cunxin, Gao Kai, Lyu Chaoliang, Zhang Kefeng. Mechanism by which nobiletin inhibits inflammatory response of BV2 microglia [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1321-1327. |
| [8] | Zheng Rongfa, Mo Weibin, Huang Peng, Chen Junji, Liang Ting, Zi Fangyu, Li Guofeng. Effects of electroacupuncture on the expression of metabolic enzymes and autophagy genes in gastrocnemius muscle tissues of exercising rats [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1127-1136. |
| [9] | Chen Yuning, Jiang Ying, Liao Xiangyu, Chen Qiongjun, Xiong Liang, Liu Yue, Liu Tong. Buqi Huoxue Compounds intervene with the expression of related factors and autophagy related proteins in a rat model of cerebral ischemia/reperfusion [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1152-1158. |
| [10] | Liu Lingyun, He Guixin, Qin Weibin, Song Hui, Zhang Liwen, Tang Weizhi, Yang Feifei, Zhu Ziyi, Ou Yangbin . Improvement of myocardial injury by traditional Chinese medicine: mitochondrial calcium homeostasis mediates macrophage autophagy and pyroptosis pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1276-1284. |
| [11] | Xu Tianjie, Fan Jiaxin, Guo Xiaoling, Jia Xiang, Zhao Xingwang, Liu kainan, Wang Qian. Metformin exerts a protective effect on articular cartilage in osteoarthritis rats by inhibiting the PI3K/AKT/mTOR pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1003-1012. |
| [12] | He Yang, Tang Buyuan, Lu Changhuai. Molecular mechanisms of ligament flavum hypertrophy: analysis based on methylation sequencing and transcriptome integration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1013-1020. |
| [13] | Yu Hui, Yang Yang, Wei Ting, Li Wenli, Luo Wenqian, Liu Bin. Gadd45b alleviates white matter damage in chronic ischemic rats by modulating astrocyte phenotype [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7797-7803. |
| [14] | Liu Xuan, Ding Yuqing, Xia Ruohan, Wang Xianwang, Hu Shujuan. Exercise prevention and treatment of insulin resistance: role and molecular mechanism of Keap1/nuclear factor erythroid2-related factor 2 signaling pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7578-7588. |
| [15] | Gong Yuehong, Wang Mengjun, Ren Hang, Zheng Hui, Sun Jiajia, Liu Junpeng, Zhang Fei, Yang Jianhua, Hu Junping. Machine learning combined with bioinformatics screening of key genes for pulmonary fibrosis associated with cellular autophagy and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7679-7689. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||