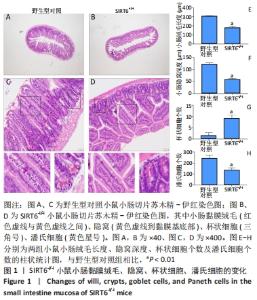
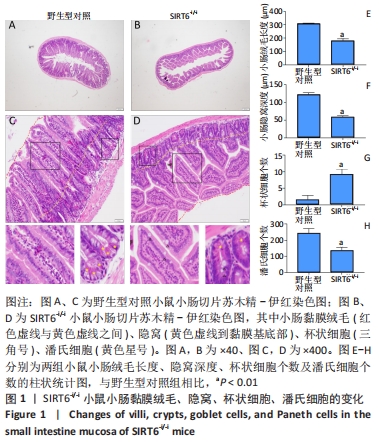
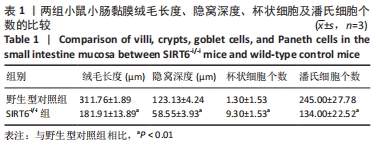
[1] 蒋智, 田子林, 胡帅, 等. NAD+代谢与衰老及其运动适应研究进展[J]. 体育科研,2021,42(5):12-19.
[2] 陈杰, 万剑华, 吴当彦, 等 SIRT在肠屏障中作用的研究进展[J]. 基础医学与临床,2018,38(1):103-106.
[3] MOSTOSLAVSKY R, CHUA KF, LOMBARD DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315-329.
[4] LIU F, BU HF, GENG H, et al. Sirtuin-6 preserves R-spondin-1 expression and increases resistance of intestinal epithelium to injury in mice. Mol Med. 2017;23:272-284.
[5] PAN H, GUAN D, LIU X, et al. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 2016; 26(2):190-205.
[6] ZHANG P, LIU Y, WANG Y, et al. SIRT6 promotes osteogenic differentiation of mesenchymal stem cells through BMP signaling. Sci Rep. 2017;7(1):10229.
[7] ETCHEGARAY JP, CHAVEZ L, HUANG Y, et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol. 2015;17(5):545-557.
[8] Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480-484.
[9] KOROTKOV A, SELUANOV A, GORBUNOVA V. Sirtuin 6: linking longevity with genome and epigenome stability. Trends Cell Biol. 2021;31(12): 994-1006.
[10] ROICHMAN A, ELHANATI S, AON MA, et al. Restoration of energy homeostasis by SIRT6 extends healthy lifespan. Nat Commun. 2021; 12(1):3208.
[11] SUN H, WU Y, FU D, et al. SIRT6 regulates osteogenic differentiation of rat bone marrow mesenchymal stem cells partially via suppressing the nuclear factor-kappaB signaling pathway. Stem Cells. 2014;32(7):1943-1955.
[12] WANG H, DIAO D, SHI Z, et al. SIRT6 Controls Hematopoietic Stem Cell Homeostasis through Epigenetic Regulation of Wnt Signaling. Cell Stem Cell. 2016;18(4):495-507.
[13] 刘芳宜. SIRT6在溃疡性结肠炎发病机制中的作用研究[D]. 北京:北京协和医学院;中国医学科学院;清华大学医学部;北京协和医学院中国医学科学院内科学(消化内科), 北京协和医学院, 2015.
[14] LU H, LIN J, XU C, et al. Cyclosporine modulates neutrophil functions via the SIRT6-HIF-1α-glycolysis axis to alleviate severe ulcerative colitis. Clin Transl Med. 2021;11(2):e334.
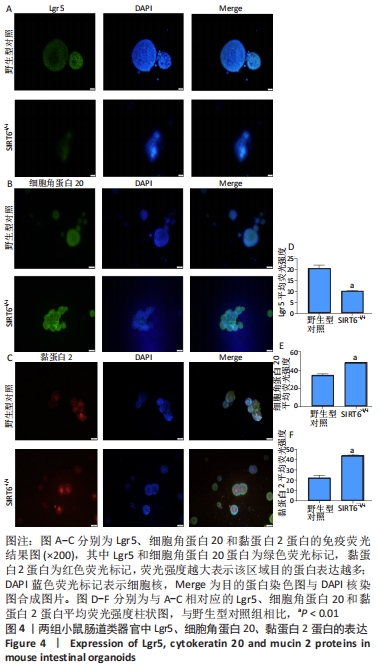
[15] BARKER N, VAN ES JH, KUIPERS J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007; 449(7165):1003-1007.
[16] CRAY P, SHEAHAN BJ, DEKANEY CM. Secretory Sorcery: Paneth Cell Control of Intestinal Repair and Homeostasis. Cell Mol Gastroenterol Hepatol. 2021;12(4):1239-1250.
[17] GEHART H, CLEVERS H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol. 2019;16(1):19-34.
[18] 夏青松,徐丽君,陈广,等. 信号分子通路网络对肠干细胞增殖和分化的调控[J]. 生理科学进展,2020,51(4):305-310.
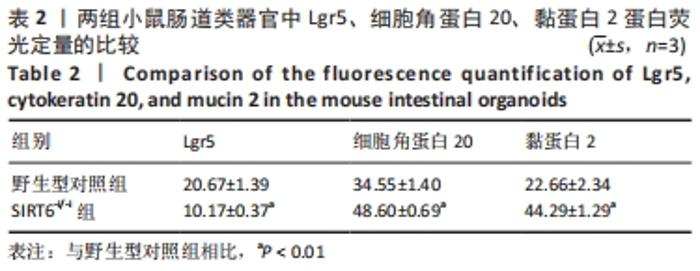
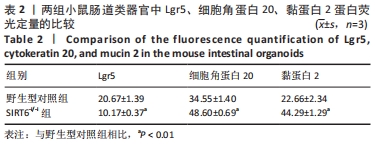
[19] 刘睿, 杨垒, 颜浩. CK20、Nm23与Ki67蛋白在结直肠癌组织中的表达及意义[J]. 中国现代普通外科进展,2020,23(10):797-800.
[20] 李冰,于岩波.肠黏液屏障在肠道中的作用[J]. 世界华人消化杂志, 2017,25(19):1764-1771.
[21] LEE SY, CHOI DW, JANG KT, et al. High Expression of Intestinal-Type Mucin (MUC2) in Intraductal Papillary Mucinous Neoplasms Coexisting With Extrapancreatic Gastrointestinal Cancers. Pancreas. 2006;32(2): 186-189.
[22] BUISINE MP, DEVISME L, DEGAND P, et al. Developmental mucin gene expression in the gastroduodenal tract and accessory digestive glands. II. Duodenum and liver, gallbladder, and pancreas. J Histochem Cytochem. 2000;48(12):1667-1676.
[23] 黄玉松, 李力. 细胞色素P450酶参与动物体内代谢的研究与展望[J]. 药物生物技术,2021,28(1):94-98.
[24] NEBERT DW. Drug metabolism. Growth signal pathways. Nature. 1990; 347(6295):709-710.
[25] STEVENS JC, HINES RN, GU C, et al. Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther. 2003; 307(2):573-582.
[26] KUNO T, HIRAYAMA-KUROGI M, ITO S, et al. Effect of Intestinal Flora on Protein Expression of Drug-Metabolizing Enzymes and Transporters in the Liver and Kidney of Germ-Free and Antibiotics-Treated Mice. Mol Pharm. 2016;13(8):2691-2701.
[27] 兰轲. 药物代谢酶CYP3A在胆汁酸宿主-肠道微生物协同代谢中的生物学功能假说[J]. 中国临床药理学杂志,2019,35(22):2923-2929.
[28] MROZ MS, LAJCZAK NK, GOGGINS BJ, et al. The bile acids, deoxycholic acid and ursodeoxycholic acid, regulate colonic epithelial wound healing. Am J Physiol Gastrointest Liver Physiol. 2018;314(3): G378-G387.
[29] SONG M, YE J, ZHANG F, et al. Chenodeoxycholic Acid (CDCA) Protects against the Lipopolysaccharide-Induced Impairment of the Intestinal Epithelial Barrier Function via the FXR-MLCK Pathway. J Agric Food Chem. 2019;67(32):8868-8874.
|