Chinese Journal of Tissue Engineering Research ›› 2022, Vol. 26 ›› Issue (29): 4672-4679.doi: 10.12307/2022.883
Previous Articles Next Articles
Exploring peripheral blood biomarkers and therapeutic drugs for osteoarthritis based on bioinformatics
Yang Wei1, Han Qingmin2
- 1The Third Clinical Medical College of Guangzhou University of Chinese Medicine, Guangzhou 510405, Guangdong Province, China; 2The Third Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou 510405, Guangdong Province, China
-
Received:2021-09-30Accepted:2021-12-11Online:2022-10-18Published:2022-03-28 -
Contact:Han Qingmin, MD, Chief physician, Professor, Doctoral supervisor, The Third Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou 510405, Guangdong Province, China -
About author:Yang Wei, MD candidate, Attending physician, The Third Clinical Medical College of Guangzhou University of Chinese Medicine, Guangzhou 510405, Guangdong Province, China -
Supported by:A Pilot Project of Clinical Collaboration between Traditional Chinese Medicine and Western Medicine for the Major and Difficult Diseases-Degenerative Osteoarthrosis (Guangdong), No. State Council Medical Affairs Office [2018]3 (to HQM)
CLC Number:
Cite this article
Yang Wei, Han Qingmin. Exploring peripheral blood biomarkers and therapeutic drugs for osteoarthritis based on bioinformatics[J]. Chinese Journal of Tissue Engineering Research, 2022, 26(29): 4672-4679.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
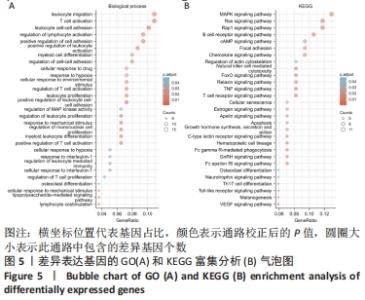
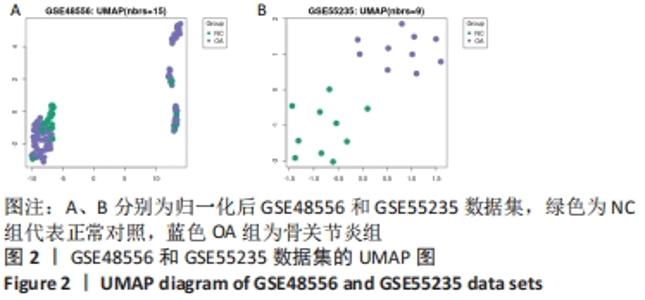
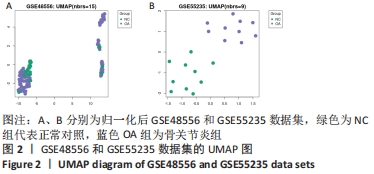
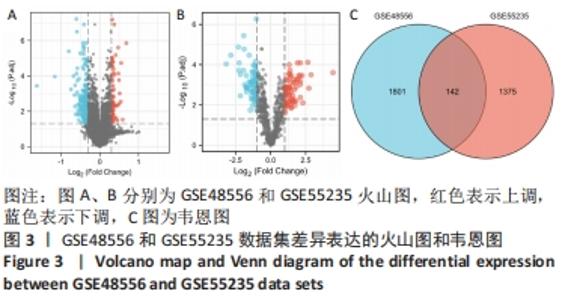
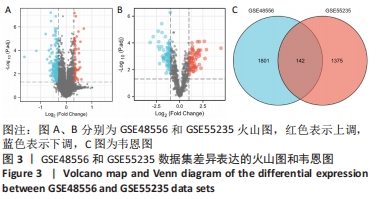
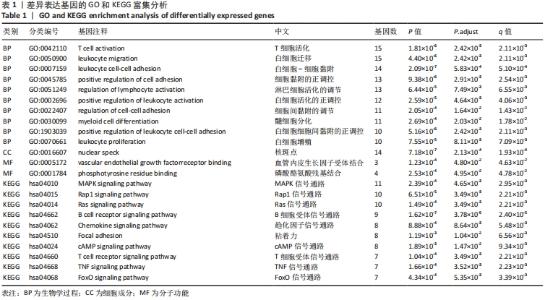
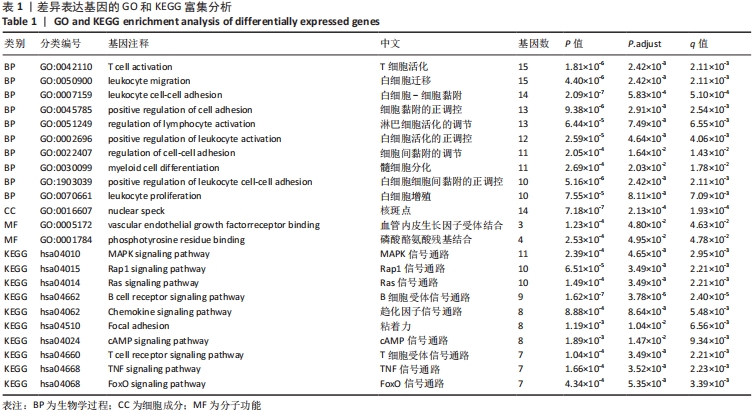
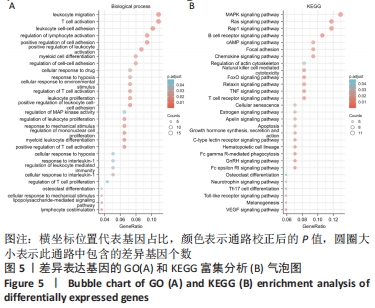
2.3 GO功能和KEGG通路富集分析 对筛选得到的共有差异表达基因进行GO和KEGG富集分析,共富集到193条,其中在满足P.adj< 0.05 & q value< 0.2条件下,BP共有61条,CC共有1条,MF共有2条,KEGG共有55条。BP主要富集在T细胞活化、白细胞迁移、白细胞-细胞黏附、细胞黏附的正调控和淋巴细胞活化的调节等条目;CC富集在细胞的核斑点;MF富集在血管内皮生长因子受体结合和磷酸酪氨酸残基结合;KEGG富集在MAPK信号通路、Rap1信号通路、Ras信号通路、B细胞受体信号通路和趋化因子信号通路等通路。在满足P.adjust< 0.05& Count≥5条件下,结合与骨关节炎的相关性展示BP和KEGG,分别有29条,表格仅展示前10条,具体见图5和表1。"
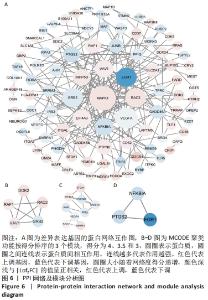
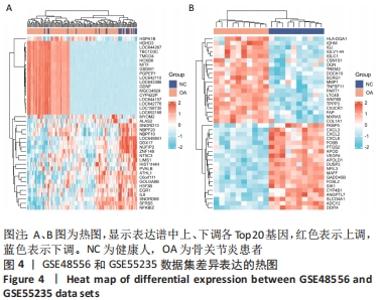
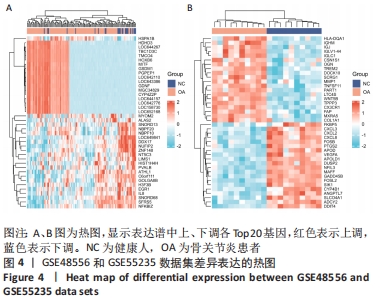
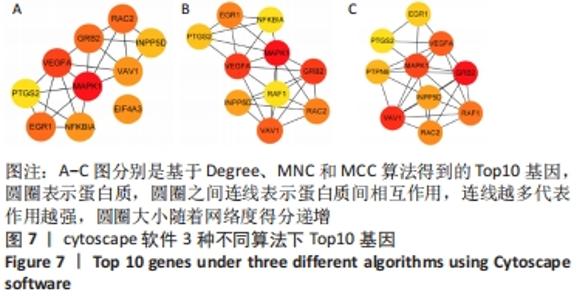
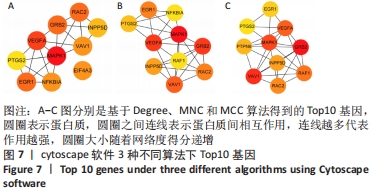
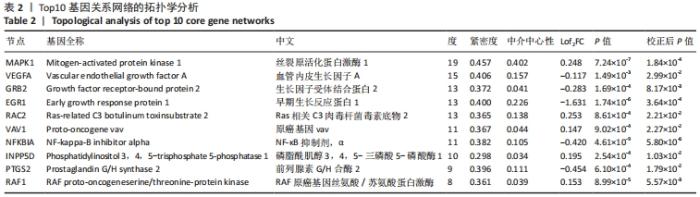
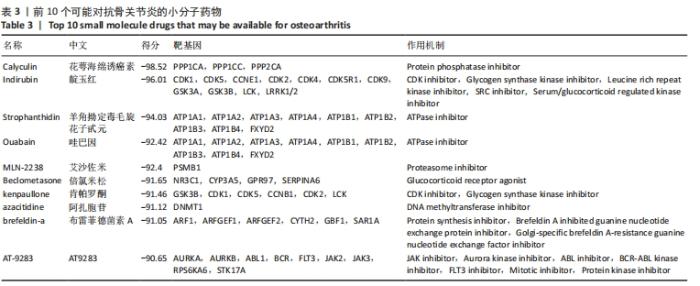
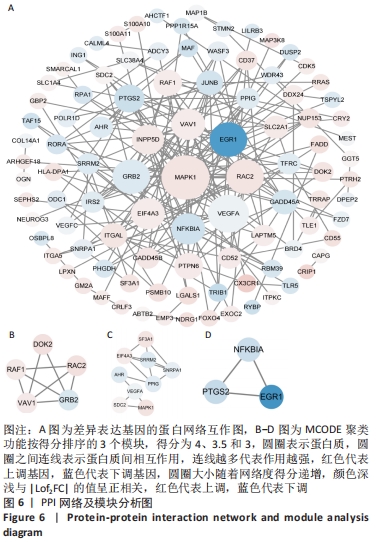
2.4 差异表达基因的PPI网络分析及Top基因 将筛选到的142个差异表达基因导入STRING在线平台,结果导入Cytoscape-v3.8.2中,得到PPI网络图,共有98个节点,184条互作关系,经MCODE筛选出4个集簇,展示得分较高的前3个集簇,具体见图6。然后使用cytohubba插件采用MNC、MCC和Degree算法筛选出连结度最高的前10个基因,再联合度(Degree)、中介中心性(Betweenness)和紧密度(Closeness)筛选Top10基因:MAPK1、VEGFA、GRB2、EGR1、RAC2、VAV1、NFKBIA、INPP5D、PTGS2和RAF1。见图7和表2。"
| [1] Macfarlane E, Seibel MJ, Zhou H. et al. Arthritis and the role of endogenous glucocorticoids. Bone Res. 2020;8:33. [2] HUNTER DJ, BIERMA-ZEINSTRA S. Osteoarthritis. Lancet. 2019;393 (10182):1745-1759. [3] Miller RE, Scanzello CR, Malfait AM. An emerging role for toll-like receptors at the neuroimmune interface in osteoarthritis. Semin Immunopathol. 2019;41(5):583-594. [4] Aubourg G, Rice SJ, Bruce-Wootton P, et al. Genetics of osteoarthritis. Osteoarthritis Cartilage. 2021;S1063-4584(21)00632-4. doi: 10.1016/j.joca.2021.03.002. [5] Ratneswaran A, Kapoor M. Osteoarthritis year in review: genetics, genomics, epigenetics. Osteoarthritis Cartilage. 2021;29(2):151-160. [6] Coryell PR, Diekman BO, Loeser RF. Mechanisms and therapeutic implications of cellular senescence in osteoarthritis. Nat Rev Rheumatol. 2021;17(1):47-57. [7] Veret D, Jorgensen C, Brondello JM. Osteoarthritis in time for senotherapeutics. Joint Bone Spine. 2021;88(2):105084. [8] Kulkarni P, Martson A, Vidya R, et al. Pathophysiological landscape of osteoarthritis. Adv Clin Chem. 2021;100:37-90. [9] Perry TA, Parkes MJ, Hodgson RJ, et al. Association between Bone marrow lesions & synovitis and symptoms in symptomatic knee osteoarthritis. Osteoarthritis Cartilage. 2020;28(3):316-323. [10] KATZ JN, ARANT KR, LOESER RF. Diagnosis and Treatment of Hip and Knee Osteoarthritis: A Review. JAMA. 2021;325(6):568-578. [11] Hunter DJ, March L, Chew M. Osteoarthritis in 2020 and beyond: a Lancet Commission. Lancet. 2020;396(10264):1711-1712. [12] Szilagyi IA, Waarsing JH, Schiphof D, et al. Towards sex-specific osteoarthritis risk models: evaluation of risk factors for knee osteoarthritis in males and females. Rheumatology (Oxford). 2021;keab378. doi: 10.1093/rheumatology/keab378. [13] SUN X, ZHEN X, HU X, et al. Osteoarthritis in the Middle-Aged and Elderly in China: Prevalence and Influencing Factors. Int J Environ Res Public Health. 2019;16(23):4701. [14] Ma L, Chhetri JK, Zhang L, et al. Cross-sectional study examining the status of intrinsic capacity decline in community-dwelling older adults in China: prevalence, associated factors and implications for clinical care. BMJ Open. 2021;11(1):e043062. [15] 孙青,梁锴,李岩.血液样本质量的评估技术进展[J].生物化学与生物物理进展,2021,48(8):938-946. [16] 周巧,刘健,宋倩,等.膝骨关节炎患者外周血单个核细胞CD36表达与氧化应激的相关性研究[J].风湿病与关节炎,2021,10(3):6-11. [17] WENDLING D, ABBAS W, GODFRIN-VALNET M, et al. Resveratrol, a sirtuin 1 activator, increases IL-6 production by peripheral blood mononuclear cells of patients with knee osteoarthritis. Clin Epigenetics. 2013;5(1):10. [18] Soyocak A, Kurt H, Ozgen M, et al. miRNA-146a, miRNA-155 and JNK expression levels in peripheral blood mononuclear cells according to grade of knee osteoarthritis. Gene. 2017;627:207-211. [19] Okuhara A, Nakasa T, Shibuya H, et al. Changes in microRNA expression in peripheral mononuclear cells according to the progression of osteoarthritis. Mod Rheumatol. 2012;22(3):446-457. [20] Ramos YF, Bos SD, Lakenberg N, et al. Genes expressed in blood link osteoarthritis with apoptotic pathways. Ann Rheum Dis. 2014;73(10): 1844-1853. [21] Feng Z, Lian KJ. Identification of genes and pathways associated with osteoarthritis by bioinformatics analyses. Eur Rev Med Pharmacol Sci. 2015;19(5):736-744. [22] Li J, Lan CN, Kong Y, et al. Identification and Analysis of Blood Gene Expression Signature for Osteoarthritis With Advanced Feature Selection Methods. Front Genet. 2018;9:246. [23] ZHANG W, QIU Q, SUN B, et al. A four-genes based diagnostic signature for osteoarthritis. Rheumatol Int. 2021;41(10):1815-1823. [24] Riyazi N, Meulenbelt I, Kroon HM, et al. Evidence for familial aggregation of hand, hip, and spine but not knee osteoarthritis in siblings with multiple joint involvement: the GARP study. Ann Rheum Dis. 2005;64(3):438-443. [25] Shi T, Shen X, Gao G. Gene Expression Profiles of Peripheral Blood Monocytes in Osteoarthritis and Analysis of Differentially Expressed Genes. Biomed Res Int. 2019;2019:4291689. [26] Cai P, Jiang T, Li B, et al. Comparison of rheumatoid arthritis (RA) and osteoarthritis (oa) based on microarray profiles of human joint fibroblast-like synoviocytes. Cell Biochem Funct. 2019;37(1):31-41. [27] SUBRAMANIAN A, NARAYAN R, CORSELLO SM, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171(6):1437-1452. [28] Enache OM, Lahr DL, Natoli TE, et al The GCTx format and cmap{Py, R, M, J} packages: resources for optimized storage and integrated traversal of annotated dense matrices. Bioinformatics. 2019; 35(8):1427-1429. [29] 袁普卫.李氏四辨思想在膝骨关节炎防治中的作用[J].中国中西医结合杂志,2021,41(7):782-783. [30] 李金明,王辉,张志毅.骨关节炎诊断生物标记物研究进展[J].中华风湿病学杂志,2020,24(9):631-634. [31] 解英,陈斌,汪虹,等.外周血单个核细胞来源的多潜能细胞的研究进展[J].中华烧伤杂志,2016,32(12):762-764. [32] 李伯阳,赵艳玲,田玉玲,等.轮状病毒感染患者外周血单个核细胞的转录组学分析[J/OL].病毒学报,2021, https://doi.org/10.13242/j.cnki.bingduxuebao.003963. [33] 郭朋朋,刘亚妮,师少军,等.器官移植受者外周血单个核细胞中免疫抑制剂药物浓度的影响因素研究进展[J].中国医院药学杂志, 2021,41(8):855-859. [34] 魏叶,刘亚妮,师少军,等.外周血单个核细胞内免疫抑制剂的浓度与实体器官移植术后不良反应相关性的研究进展[J].中国医院药学杂志,2020,40(8):946-951. [35] Ponchel F, Burska AN, Hensor EM, et al. Changes in peripheral blood immune cell composition in osteoarthritis. Osteoarthritis Cartilage. 2015;23(11):1870-1878. [36] Moradi B, Rosshirt N, Tripel E, et al. Unicompartmental and bicompartmental knee osteoarthritis show different patterns of mononuclear cell infiltration and cytokine release in the affected joints. Clin Exp Immunol. 2015;180(1):143-154. [37] 王猛,陈敬东,王佳蔚. 膝关节骨关节炎外周血标志物变化的临床研究[J].重庆医科大学学报,2021,46(3):306-310. [38] Wu CL, Harasymowicz NS, Klimak MA, et al. The role of macrophages in osteoarthritis and cartilage repair. Osteoarthritis Cartilage. 2020;28(5):544-554. [39] Thomson A, Hilkens CMU. Synovial Macrophages in Osteoarthritis: The Key to Understanding Pathogenesis? Front Immunol. 2021;12: 678757. [40] Zhang H, Cai D, Bai X. Macrophages regulate the progression of osteoarthritis. Osteoarthritis Cartilage. 2020;28(5):555-561. [41] Kooistra MR, Dubé N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120(1):17-22. [42] Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol. 2009;21(5):684-693. [43] Xia Z, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci. 2005;6(4):267-276. [44] Doebele RC, Schulze-Hoepfner FT, Hong J, et al. A novel interplay between Epac/Rap1 and mitogen-activated protein kinase kinase 5/extracellular signal-regulated kinase 5 (MEK5/ERK5) regulates thrombospondin to control angiogenesis. Blood. 2009;114(20): 4592-4600. [45] Minato N, Kometani K, Hattori M. Regulation of Immune Responses and Hematopoiesis by the Rap1 Signal. 2007:229-264. [46] Breckler M, Berthouze M, Laurent AC, et al. Rap-linked cAMP signaling Epac proteins: Compartmentation, functioning and disease implications. Cellular Signalling. 2011;23(8):1257-1266. [47] 于雅丽,孔奕翼,叶静,等.基于综合生物信息学筛选骨关节炎潜在生物标记物的研究[J].中华创伤骨科杂志,2021,23(1):75-80. [48] STRZELECKA-KILISZEK A, MEBAREK S, ROSZKOWSKA M, et al. Functions of Rho family of small GTPases and Rho-associated coiled-coil kinases in bone cells during differentiation and mineralization. Biochim Biophys Acta Gen Subj. 2017;1861(5 Pt A):1009-1023. [49] HOU SM, HOU CH, LIU JF. CX3CL1 promotes MMP-3 production via the CX3CR1, c-Raf, MEK, ERK, and NF-κB signaling pathway in osteoarthritis synovial fibroblasts. Arthritis Res Ther. 2017;19(1):282. [50] Stiffel VM, Thomas A, Rundle CH, et al. The EphA4 Signaling is Anti‑catabolic in Synoviocytes but Pro‑anabolic in Articular Chondrocytes. Calcif Tissue Int. 2020;107(6):576-592. [51] HAMILTON JL, NAGAO M, LEVINE BR, et al. Targeting VEGF and its Receptors for the Treatment of Osteoarthritis and Associated Pain. J Bone Miner Res. 2016;31(5):911-924. [52] GUAN M, ZHU Y, LIAO B, et al. Low-intensity pulsed ultrasound inhibits VEGFA expression in chondrocytes and protects against cartilage degeneration in experimental osteoarthritis. FEBS Open Bio. 2020;10(3):434-443. [53] Xu K, Gao Y, Yang L, et al. Magnolin exhibits anti-inflammatory effects on chondrocytes via the NF-kappaB pathway for attenuating anterior cruciate ligament transection-induced osteoarthritis. Connect Tissue Res. 2021;62(4):475-484. [54] LI Z, WANG H, WEI J, et al. Indirubin exerts anticancer effects on human glioma cells by inducing apoptosis and autophagy. AMB Express. 2020; 10(1):171. [55] LAI J, LIU Y, LIU C, et al. Indirubin Inhibits LPS-Induced Inflammation via TLR4 Abrogation Mediated by the NF-kB and MAPK Signaling Pathways. Inflammation. 2017;40(1):1-12. |
| [1] | LIU Danni, SUN Guanghua, ZHOU Guijuan, LIU Hongya, ZHOU Jun, TAN Jinqu, HUANG Xiarong, PENG Ting, FENG Wei-bin, LUO Fu. Effect of electroacupuncture on apoptosis of neurons in cerebral cortex of rats with cerebral ischemia-reperfusion injury at "Shuigou" and "Baihui" points [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(在线): 1-6. |
| [2] | Jin Tao, Liu Lin, Zhu Xiaoyan, Shi Yucong, Niu Jianxiong, Zhang Tongtong, Wu Shujin, Yang Qingshan. Osteoarthritis and mitochondrial abnormalities [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1452-1458. |
| [3] | Zhang Jichao, Dong Yuefu, Mou Zhifang, Zhang Zhen, Li Bingyan, Xu Xiangjun, Li Jiayi, Ren Meng, Dong Wanpeng. Finite element analysis of biomechanical changes in the osteoarthritis knee joint in different gait flexion angles [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1357-1361. |
| [4] | Wang Baojuan, Zheng Shuguang, Zhang Qi, Li Tianyang. Miao medicine fumigation can delay extracellular matrix destruction in a rabbit model of knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(8): 1180-1186. |
| [5] | Xiang Xinjian, Liu Fang, Wu Liangliang, Jia Daping, Tao Yue, Zhao Zhengnan, Zhao Yu. High-dose vitamin C promotes the survival of autologous fat transplantation in rats [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(8): 1242-1246. |
| [6] | Wen Dandan, Li Qiang, Shen Caiqi, Ji Zhe, Jin Peisheng. Nocardia rubra cell wall skeleton for extemal use improves the viability of adipogenic mesenchymal stem cells and promotes diabetes wound repair [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1038-1044. |
| [7] | Zhang Yujie, Yang Jiandong, Cai Jun, Zhu Shoulei, Tian Yuan. Mechanism by which allicin inhibits proliferation and promotes apoptosis of rat vascular endothelial cells [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1080-1084. |
| [8] | Liu Dongcheng, Zhao Jijun, Zhou Zihong, Wu Zhaofeng, Yu Yinghao, Chen Yuhao, Feng Dehong. Comparison of different reference methods for force line correction in open wedge high tibial osteotomy [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(6): 827-831. |
| [9] | Zhou Jianguo, Liu Shiwei, Yuan Changhong, Bi Shengrong, Yang Guoping, Hu Weiquan, Liu Hui, Qian Rui. Total knee arthroplasty with posterior cruciate ligament retaining prosthesis in the treatment of knee osteoarthritis with knee valgus deformity [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(6): 892-897. |
| [10] | Xu Lei, Han Xiaoqiang, Zhang Jintao, Sun Haibiao. Hyaluronic acid around articular chondrocytes: production, transformation and function characteristics [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 768-773. |
| [11] | Liu Yiyi, Qiu Junqiang, Yi Longyan, Zhou Cailiang. Effect of resistance training on interleukin-6 and C-reactive protein in middle-age and elderly people: a Meta-analysis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 804-812. |
| [12] | He Junjun, Huang Zeling, Hong Zhenqiang. Interventional effect of Yanghe Decoction on synovial inflammation in a rabbit model of early knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 694-699. |
| [13] | Lin Xuchen, Zhu Hainian, Wang Zengshun, Qi Tengmin, Liu Limin, Suonan Angxiu. Effect of xanthohumol on inflammatory factors and articular cartilage in a mouse mode of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 676-681. |
| [14] | Deng Shuang, Pu Rui, Chen Ziyang, Zhang Jianchao, Yuan Lingyan . Effects of exercise preconditioning on myocardial protection and apoptosis in a mouse model of myocardial remodeling due to early stress overload [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 717-723. |
| [15] | Zhang Tong, Cai Jinchi, Yuan Zhifa, Zhao Haiyan, Han Xingwen, Wang Wenji. Hyaluronic acid-based composite hydrogel in cartilage injury caused by osteoarthritis: application and mechanism [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 617-625. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||