Chinese Journal of Tissue Engineering Research ›› 2022, Vol. 26 ›› Issue (23): 3706-3713.doi: 10.12307/2022.671
Previous Articles Next Articles
Bioinformatics analysis of gene expression profile of peripheral blood lymphocytes in patients with osteoarthritis
Yang Wei1, Yuan Puwei2, Du Longlong2, Li Xuefeng2, Gao Qimeng2, Han Qingmin3
- 1The Third Clinical Medical College of Guangzhou University of Chinese Medicine, Guangzhou 510405, Guangdong Province, China; 2Shaanxi University of Chinese Medicine, Xianyang 712046, Shaanxi Province, China; 3the Third Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou 510405, Guangdong Province, China
-
Received:2021-08-07Accepted:2021-09-08Online:2022-08-18Published:2022-02-23 -
Contact:Han Qingmin, MD, Chief physician, Professor, Doctoral supervisor, the Third Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou 510405, Guangdong Province, China -
About author:Yang Wei, MD candidate, Attending physician, the Third Clinical Medical College of Guangzhou University of Chinese Medicine, Guangzhou 510405, Guangdong Province, China -
Supported by:the Pilot Project of Clinical Collaboration between Traditional Chinese Medicine and Western Medicine for the Major and Difficult Diseases-Degenerative Osteoarthritis in Guangdong Province approved by State Council Medical Affairs Office, No. [2018]3 (to HQM)
CLC Number:
Cite this article
Yang Wei, Yuan Puwei, Du Longlong, Li Xuefeng, Gao Qimeng, Han Qingmin. Bioinformatics analysis of gene expression profile of peripheral blood lymphocytes in patients with osteoarthritis[J]. Chinese Journal of Tissue Engineering Research, 2022, 26(23): 3706-3713.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
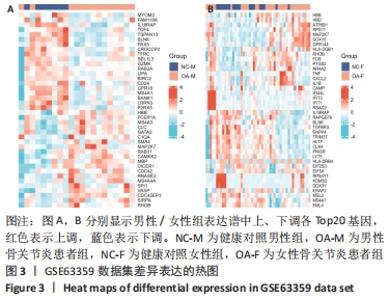
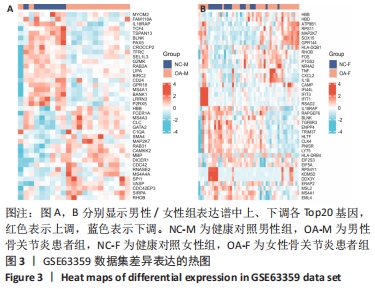
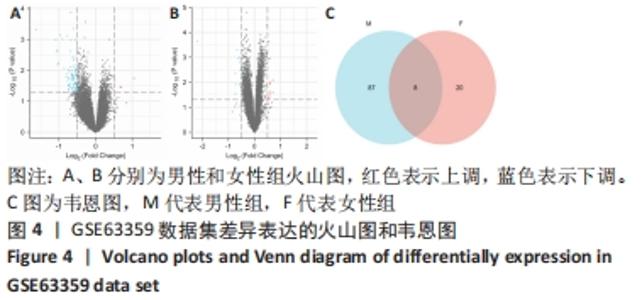
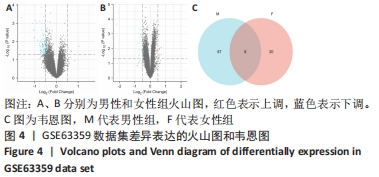
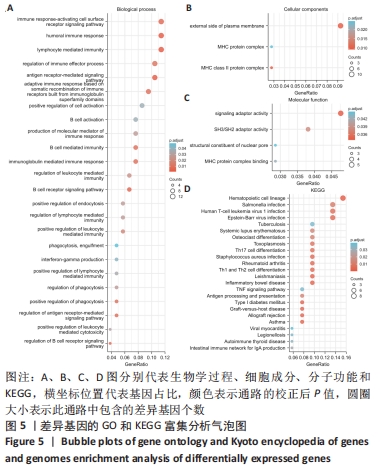
2.3 差异表达基因的GO功能和KEGG通路富集分析 对筛选得到的所有差异表达基因进行GO和KEGG富集分析,共富集到88条,其中在满足P.adj < 0.05 & q value < 0.05条件下,生物学过程共有24条,CC共有3条,MF共有4条,KEGG共有24条。BP主要富集在“淋巴细胞介导的免疫”“体液免疫反应”“抗原受体介导的信号通路”“B细胞受体信号通路”“免疫球蛋白介导的免疫反应”和“吞噬作用的正调控”等24条生物学过程上;CC富集在“质膜外侧”“MHC Ⅱ类蛋白质复合物”“MHC蛋白复合物”等细胞成分中;MF富集在“信号适配器活动”“SH3/SH2 适配器活动”“MHC 蛋白复合物结合”“核孔结构成分”等分子功能中;KEGG富集在造血细胞谱系、哮喘、炎症性肠病、同种异体排斥、利什曼病、移植物抗宿主病、1型糖尿病、Th1和Th2细胞分化、Th17细胞分化、破骨细胞分化和TNF信号通路等通路上。具体见表2和图5。"
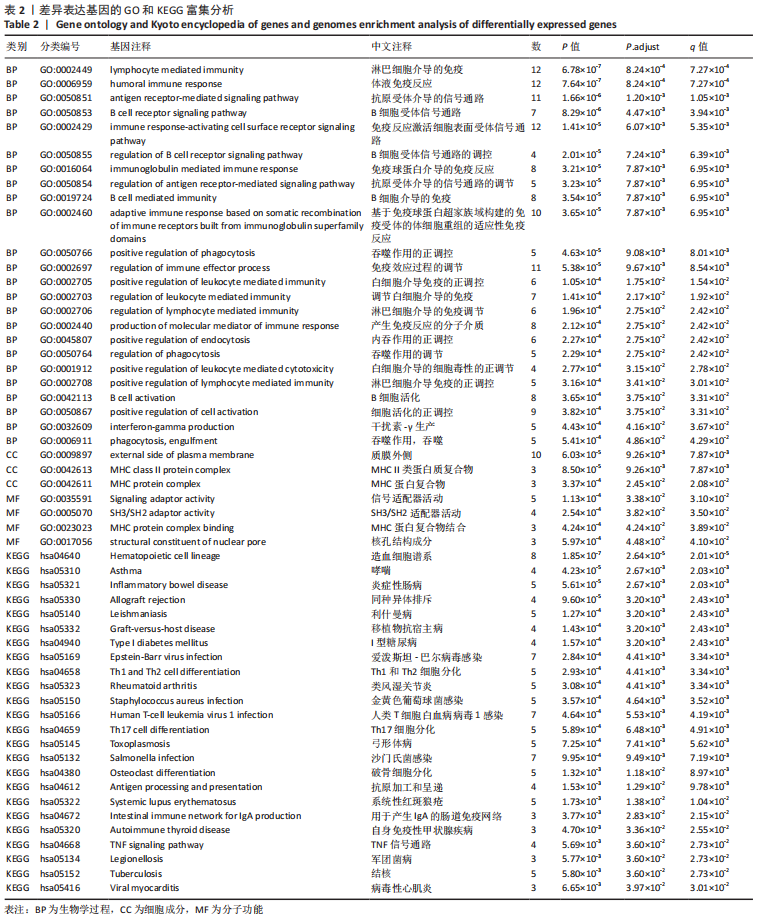
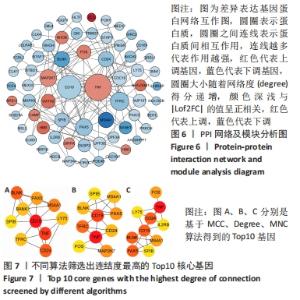
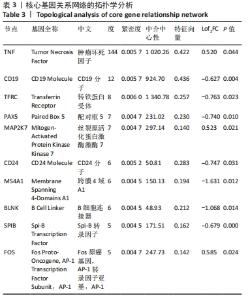
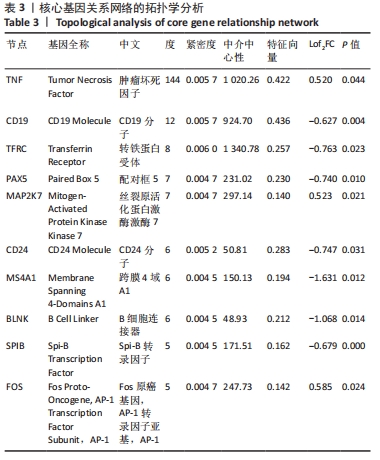
2.4 PPI网络分析及模块分析结果 将筛选到的差异表达基因导入STRING在线平台,其结果导入Cytoscape-v 3.8.2中,得到PPI网络图,共有66个节点,105条互作关系,见图6;经MCODE筛选出4个集簇,展示得分均较低。然后使用cytohubba插件采用MCC、Degree和MNC算法筛选出连结度最高的前10个基因,再联合centiscape插件中度(Degree)、中介中心性(Betweenness)、特征向量(Eigenvector)和紧密度(Closeness)等参数以及对应的P值和log2FC值筛选核心基因,之后将Degree法和MCC法以及Degree法和MNC法筛选的前10基因分别取交集,最后两个交集中共同有8个差异基因为:TNF(肿瘤坏死因子)、CD19、TFRC(转铁蛋白受体)、PAX5(配对框5)、MAP2K7(丝裂原活化蛋白激酶激酶7)、CD24、MS4A1(跨膜4域A1)和BLNK(B细胞连接器),见图7和表3。"
| [1] ARDEN NK, PERRY TA, BANNURU RR, et al. Non-surgical management of knee osteoarthritis: comparison of ESCEO and OARSI 2019 guidelines. Nat Rev Rheumatol. 2021;17(1):59-66. [2] VOS T, LIM SS, ABBAFATI C, et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020; 396(10258):1204-1222. [3] MA L, CHHETRI JK, ZHANG L, et al. Cross-sectional study examining the status of intrinsic capacity decline in community-dwelling older adults in China: prevalence, associated factors and implications for clinical care. BMJ Open. 2021;11(1):e043062. [4] WANG Y, NGUYEN U DT, LANE NE, et al. Knee Osteoarthritis, Potential Mediators, and Risk of All-Cause Mortality: Data From the Osteoarthritis Initiative. Arthritis Care Res (Hoboken). 2021;73(4):566-573. [5] LO J, CHAN L, FLYNN S. A Systematic Review of the Incidence, Prevalence, Costs, and Activity and Work Limitations of Amputation, Osteoarthritis, Rheumatoid Arthritis, Back Pain, Multiple Sclerosis, Spinal Cord Injury, Stroke, and Traumatic Brain Injury in the United States: A 2019 Update. Arch Phys Med Rehabil. 2021;102(1):115-131. [6] SAFIRI S, KOLAHI A A, CROSS M, et al. Prevalence, Deaths, and Disability-Adjusted Life Years Due to Musculoskeletal Disorders for 195 Countries and Territories 1990-2017. Arthritis Rheumatol. 2021;73(4):702-714. [7] HUNTER DJ, MARCH L, CHEW M. Osteoarthritis in 2020 and beyond: a Lancet Commission. Lancet. 2020;396(10264):1711-1712. [8] ALAHDAL M, ZHANG H, HUANG R, et al. Potential efficacy of dendritic cell immunomodulation in the treatment of osteoarthritis. Rheumatology (Oxford). 2021;60(2):507-517. [9] KULKARNI P, MARTSON A, VIDYA R, et al. Pathophysiological landscape of osteoarthritis. Adv Clin Chem. 2021;100:37-90. [10] 陈先维,陆向东,王永峰,等.骨关节炎患者单核细胞/淋巴细胞比值、红细胞压积与疾病活动度的相关性[J].昆明医科大学学报, 2019,40(6):76-79. [11] 赵广雷,王进,夏军,等. NLR、PLR及LMR在评估骨关节炎严重程度分级的应用价值[J].中国骨与关节损伤杂志,2019,34(10):1089-1091. [12] CAI C, ZHANG R, XU X, et al. Diagnostic values of NLR and miR-141 in patients with osteoarthritis and their association with severity of knee osteoarthritis. Exp Ther Med. 2021;21(1):74. [13] 王猛,陈敬东,王佳蔚. 膝关节骨关节炎外周血标志物变化的临床研究[J].重庆医科大学学报,2021,46(3):306-310. [14] FENG Z, LIAN KJ. Identification of genes and pathways associated with osteoarthritis by bioinformatics analyses. Eur Rev Med Pharmacol Sci. 2015;19(5):736-744. [15] LI J, LAN CN, KONG Y, et al. Identification and Analysis of Blood Gene Expression Signature for Osteoarthritis With Advanced Feature Selection Methods. Front Genet. 2018;9:246. [16] 于雅丽,孔奕翼,叶静,等.基于综合生物信息学筛选骨关节炎潜在生物标记物的研究[J].中华创伤骨科杂志,2021,23(1):75-80. [17] CAI P, JIANG T, LI B, et al. Comparison of rheumatoid arthritis (RA) and osteoarthritis (OA) based on microarray profiles of human joint fibroblast-like synoviocytes . Cell Biochem Funct. 2019;37(1):31-41. [18] GRIFFIN TM, SCANZELLO CR. Innate inflammation and synovial macrophages in osteoarthritis pathophysiology. Clin Exp Rheumatol. 2019;37 Suppl.120(5):57-63. [19] VAN DEN BOSCH MHJ. Osteoarthritis year in review 2020: biology. Osteoarthritis Cartilage. 2021;29(2):143-150. [20] MILLER RJ, MALFAIT AM, MILLER RE. The innate immune response as a mediator of osteoarthritis pain. Osteoarthritis Cartilage. 2020; 28(5):562-571. [21] WU CL, HARASYMOWICZ NS, KLIMAK MA, et al. The role of macrophages in osteoarthritis and cartilage repair. Osteoarthritis Cartilage. 2020;28(5):544-554. [22] ZHANG H, CAI D, BAI X. Macrophages regulate the progression of osteoarthritis. Osteoarthritis Cartilage. 2020;28(5):555-561. [23] HARRELL CR, MARKOVIC BS, FELLABAUM C, et al. Mesenchymal stem cell-based therapy of osteoarthritis: Current knowledge and future perspectives. Biomed Pharmacother. 2019;109:2318-2326. [24] MORADI B, ROSSHIRT N, TRIPEL E, et al. Unicompartmental and bicompartmental knee osteoarthritis show different patterns of mononuclear cell infiltration and cytokine release in the affected joints. Clin Exp Immunol. 2015;180(1):143-154. [25] ZHU W, ZHANG X, JIANG Y, et al. Alterations in peripheral T cell and B cell subsets in patients with osteoarthritis. Clin Rheumatol. 2020; 39(2):523-532. [26] 李金明,王辉,张志毅.骨关节炎诊断生物标记物研究进展[J].中华风湿病学杂志,2020,24(9):631-634. [27] 孙青,梁锴,李岩.血液样本质量的评估技术进展[J].生物化学与生物物理进展,2021. https://kns.cnki.net/kcms/detail/11.2161.Q.20210224.0848.002.html. [28] HUANG G, HUA S, YANG T, et al. Platelet‑rich plasma shows beneficial effects for patients with knee osteoarthritis by suppressing inflammatory factors. Exp Ther Med. 2018;15(3):3096-3102. [29] KUROKI K, WILLIAMS N, COOK J L. Histologic evidence for a humoral immune response in synovitis associated with cranial cruciate ligament disease in dogs. Veterinary Surgery. 2021;50(5):1032-1041. [30] XIE X, VAN DELFT MAM, SHUWEIHDI F, et al. Auto-antibodies to post-translationally modified proteins in osteoarthritis. Osteoarthritis Cartilage. 2021;29(6):924-933. [31] 谢锦伟,黄泽宇,裴福兴.固有免疫系统在骨关节炎发病机制中的作用及研究进展[J].中国修复与重建外科杂志,2019,33(3):370-376. [32] JIANG Y, MISHIMA H, SAKAI S, et al. Gene expression analysis of major lineage-defining factors in human bone marrow cells: effect of aging, gender, and age-related disorders. J Orthop Res. 2008;26(7):910-917. [33] KRIEGOVA E, MANUKYAN G, MIKULKOVA Z, et al. Gender-related differences observed among immune cells in synovial fluid in knee osteoarthritis. Osteoarthritis Cartilage. 2018;26(9):1247-1256. [34] GUILLEN FAJARDO R, OTERO FARINA F, MOSQUERA REY A, et al. Relationship Between the Dynamics of Telomere Loss in Peripheral Blood Leukocytes From Knee Osteoarthritis Patients and Mitochondrial DNA Haplogroups. J Rheumatol. 2021. doi: 10.3899/jrheum.201316. [35] 陈龑,薛艳,蒋鼎,等. T淋巴细胞在骨关节炎中作用的研究进展[J].医学综述,2021,27(5):833-838. [36] PENATTI A, FACCIOTTI F, DE MATTEIS R, et al. Differences in serum and synovial CD4+ T cells and cytokine profiles to stratify patients with inflammatory osteoarthritis and rheumatoid arthritis. Arthritis Res Ther. 2017;19(1):103. [37] SAE-JUNG T, SENGPRASERT P, APINUN J, et al. Functional and T Cell Receptor Repertoire Analyses of Peripheral Blood and Infrapatellar Fat Pad T Cells in Knee Osteoarthritis. J Rheumatol. 2019;46(3):309-317. [38] ZHANG H, SHI N, DIAO Z, et al. Therapeutic potential of TNFalpha inhibitors in chronic inflammatory disorders: Past and future. Genes Dis. 2021;8(1):38-47. [39] ZHANG Z, LI M, MA X, et al. GADD45beta-I attenuates oxidative stress and apoptosis via Sirt3-mediated inhibition of ER stress in osteoarthritis chondrocytes. Chem Biol Interact. 2018;296:76-82. [40] GUO X, XU T, ZHENG J, et al. Accumulation of synovial fluid CD19(+)CD24(hi)CD27(+) B cells was associated with bone destruction in rheumatoid arthritis. Sci Rep. 2020;10(1):14386. [41] KHOLODNYUK I, KADISA A, SVIRSKIS S, et al. Proportion of the CD19-Positive and CD19-Negative Lymphocytes and Monocytes within the Peripheral Blood Mononuclear Cell Set is Characteristic for Rheumatoid Arthritis. Medicina (Kaunas). 2019;55(10):630. [42] SIMÃO M, GAVAIA PJ, CAMACHO A, et al. Intracellular iron uptake is favored in Hfe-KO mouse primary chondrocytes mimicking an osteoarthritis-related phenotype. Biofactors. 2019;45(4):583-597. [43] BURTON LH, RADAKOVICH LB, MAROLF AJ, et al. Systemic iron overload exacerbates osteoarthritis in the strain 13 guinea pig. Osteoarthritis Cartilage. 2020;28(9):1265-1275. [44] LUO H, ZHANG R. Icariin enhances cell survival in lipopolysaccharide-induced synoviocytes by suppressing ferroptosis via the Xc-/GPX4 axis. Exp Ther Med. 2021;21(1):72. [45] JI Q, QI D, XU X, et al. Cryptotanshinone Protects Cartilage against Developing Osteoarthritis through the miR-106a-5p/GLIS3 Axis. Mol Ther Nucleic Acids. 2018;11:170-179. [46] LEE J, SMERIGLIO P, DRAGOO J, et al. CD24 enrichment protects while its loss increases susceptibility of juvenile chondrocytes towards inflammation. Arthritis Res Ther. 2016;18(1):292. [47] GRANDI FC, BASKAR R, SMERIGLIO P, et al. Single-cell mass cytometry reveals cross-talk between inflammation-dampening and inflammation-amplifying cells in osteoarthritic cartilage. Sci Adv. 2020; 6(11):eaay5352. [48] GEURTS J, PATEL A, HIRSCHMANN M T, et al. Elevated marrow inflammatory cells and osteoclasts in subchondral osteosclerosis in human knee osteoarthritis. J Orthop Res. 2016;34(2):262-269. |
| [1] | LIU Danni, SUN Guanghua, ZHOU Guijuan, LIU Hongya, ZHOU Jun, TAN Jinqu, HUANG Xiarong, PENG Ting, FENG Wei-bin, LUO Fu. Effect of electroacupuncture on apoptosis of neurons in cerebral cortex of rats with cerebral ischemia-reperfusion injury at "Shuigou" and "Baihui" points [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(在线): 1-6. |
| [2] | Jin Tao, Liu Lin, Zhu Xiaoyan, Shi Yucong, Niu Jianxiong, Zhang Tongtong, Wu Shujin, Yang Qingshan. Osteoarthritis and mitochondrial abnormalities [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1452-1458. |
| [3] | Zhang Jichao, Dong Yuefu, Mou Zhifang, Zhang Zhen, Li Bingyan, Xu Xiangjun, Li Jiayi, Ren Meng, Dong Wanpeng. Finite element analysis of biomechanical changes in the osteoarthritis knee joint in different gait flexion angles [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1357-1361. |
| [4] | Wang Baojuan, Zheng Shuguang, Zhang Qi, Li Tianyang. Miao medicine fumigation can delay extracellular matrix destruction in a rabbit model of knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(8): 1180-1186. |
| [5] | Xiang Xinjian, Liu Fang, Wu Liangliang, Jia Daping, Tao Yue, Zhao Zhengnan, Zhao Yu. High-dose vitamin C promotes the survival of autologous fat transplantation in rats [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(8): 1242-1246. |
| [6] | Wen Dandan, Li Qiang, Shen Caiqi, Ji Zhe, Jin Peisheng. Nocardia rubra cell wall skeleton for extemal use improves the viability of adipogenic mesenchymal stem cells and promotes diabetes wound repair [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1038-1044. |
| [7] | Zhang Yujie, Yang Jiandong, Cai Jun, Zhu Shoulei, Tian Yuan. Mechanism by which allicin inhibits proliferation and promotes apoptosis of rat vascular endothelial cells [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1080-1084. |
| [8] | Liu Dongcheng, Zhao Jijun, Zhou Zihong, Wu Zhaofeng, Yu Yinghao, Chen Yuhao, Feng Dehong. Comparison of different reference methods for force line correction in open wedge high tibial osteotomy [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(6): 827-831. |
| [9] | Zhou Jianguo, Liu Shiwei, Yuan Changhong, Bi Shengrong, Yang Guoping, Hu Weiquan, Liu Hui, Qian Rui. Total knee arthroplasty with posterior cruciate ligament retaining prosthesis in the treatment of knee osteoarthritis with knee valgus deformity [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(6): 892-897. |
| [10] | Liu Yiyi, Qiu Junqiang, Yi Longyan, Zhou Cailiang. Effect of resistance training on interleukin-6 and C-reactive protein in middle-age and elderly people: a Meta-analysis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 804-812. |
| [11] | He Junjun, Huang Zeling, Hong Zhenqiang. Interventional effect of Yanghe Decoction on synovial inflammation in a rabbit model of early knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 694-699. |
| [12] | Lin Xuchen, Zhu Hainian, Wang Zengshun, Qi Tengmin, Liu Limin, Suonan Angxiu. Effect of xanthohumol on inflammatory factors and articular cartilage in a mouse mode of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 676-681. |
| [13] | Deng Shuang, Pu Rui, Chen Ziyang, Zhang Jianchao, Yuan Lingyan . Effects of exercise preconditioning on myocardial protection and apoptosis in a mouse model of myocardial remodeling due to early stress overload [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 717-723. |
| [14] | Xu Lei, Han Xiaoqiang, Zhang Jintao, Sun Haibiao. Hyaluronic acid around articular chondrocytes: production, transformation and function characteristics [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 768-773. |
| [15] | Zhang Tong, Cai Jinchi, Yuan Zhifa, Zhao Haiyan, Han Xingwen, Wang Wenji. Hyaluronic acid-based composite hydrogel in cartilage injury caused by osteoarthritis: application and mechanism [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 617-625. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||