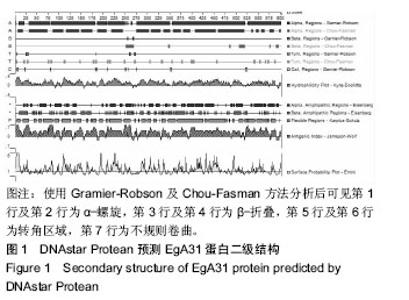
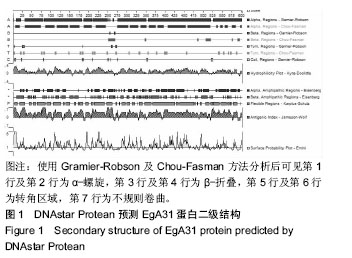
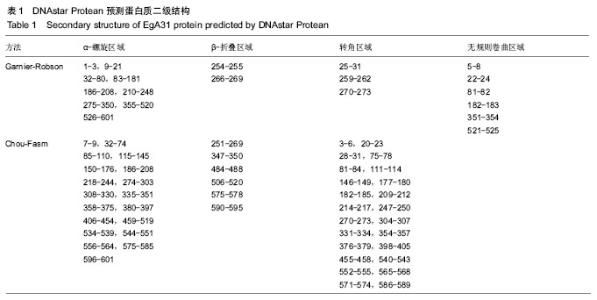
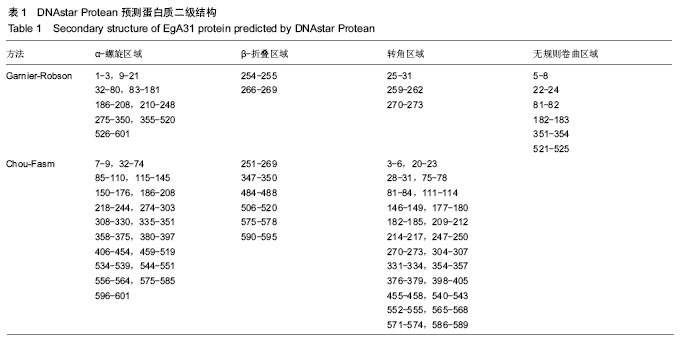
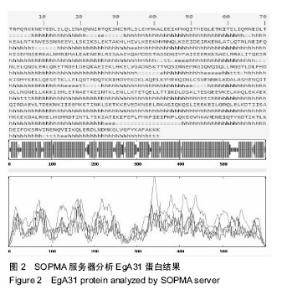
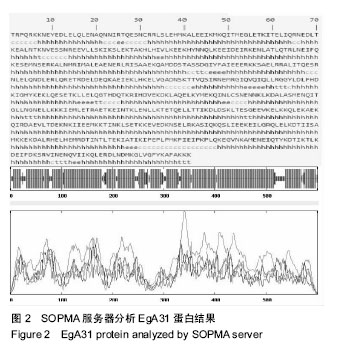
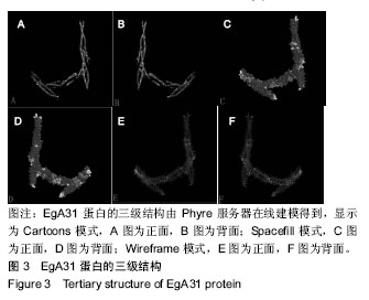
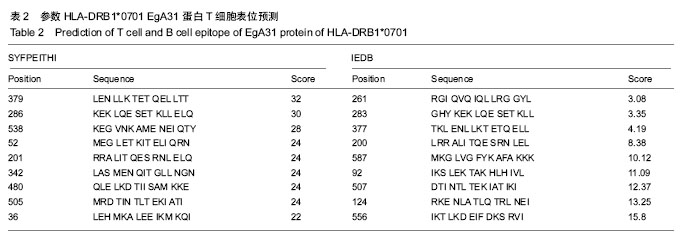
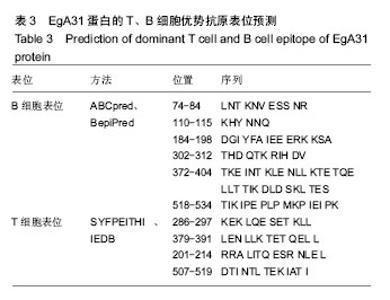
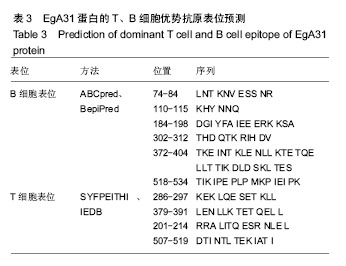
| [1] 贾海英,丁剑冰,傅玉才.细粒棘球绦虫EgA31疫苗的分子生物学特征[J].中国病原生物学杂志,2008,(7):552-555.[2] 李玉娇,王晶,赵慧,等.细粒棘球绦虫Eg95抗原表位的生物信息学预测[J].中国人兽共患病学报,2011,27(10):892-896,900.[3] Zhang FB,Lu XB,Guo N,et al.The prediction of T- and B-combined epitope of Ag85B antigen of Mycobacterium tuberculosis.Int J Clin Exp Med.2016; 9: 1408-1421[4] Fu Y, Martinez C, Chalar C,et al. A new potent antigen from Echinococcus granulosus associated with muscles and tegument.Mol Biochem Parasitol.1999;102(1):43-52.[5] 李玉娇,杨晶,赵慧,等.细粒棘球绦虫EgA31重组蛋白的抗原表位分析预测[J].中国寄生虫学与寄生虫病杂志,2012,30(1):78-80.[6] Pan Pang, Fengbo Zhang, Bin Jia, et al. Bioinformatics analysis of T- and B-combined epitopes of OMP31 protein of Brucella melitensis in Xinjiang, China.Int J Clin Exp Med.2017; 10(9):13320-13330.[7] Gasteiger E,Hoogland C,Gattiker A,et al.Protein Identification and Analysis Tools on the ExPASy Server.The Proteomics Protocols Handbook,Humana Press.2005:571-607 [8] Chen H,Gu F,Liu F.Predicting protein secondary structure using continuous wavelet transform and Chou-Fasman method. Conf Proc IEEE Eng Med Biol Soc.2005,3: 2603-2606.[9] Deléage G. ALIGNSEC: viewing protein secondary structure predictions within large multiple sequence alignments. Bioinformatics. 2017;33(24):3991-3992. [10] Kelley LA,Mezulis S,Yates CM,et al.The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015; 10(6):845-858.[11] Pikora M,Gieldon A.RASMOL AB - new functionalities in the program for structure analysis.Acta Biochim Pol.2015;62(3): 629-631.[12] Schuler MM,Nastke MD,Stevanovik? S.SYFPEITHI: database for searching and T-cell epitope prediction. Methods Mol Biol. 2007;409:75-93.[13] Karosiene E,Lundegaard C,Lund O,et al.NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics.2012;64(3):177-186.[14] Saha S,Raghava GP.Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins.2006; 65(1):40-48.[15] Larsen JE, Lund O, Nielsen M.Jens Erik Pontoppidan Larsen, Ole Lund and Morten Nielsen. Improved method for predicting linear B-cell epitopes.Immunome Res.2006;2:2.[16] Keong B,Wilkie B,Sutherland T,et al.Hepatic cystic echinococcosis in Australia: an update on diagnosis and management.ANZ J Surg.2017 Oct 11.[Epub ahead of print][17] Craig PS,Hegglin D,Lightowlers MW,et al.Echinococcosis: Control and Prevention.Adv Parasitol.2017;96:155-158.[18] 鲍佳春,袁凤来,陆伟国.包虫病治疗进展[J].中国血吸虫病防治杂,2010,22(2):197-199. [19] Grubor NM,Jovanova-Nesic KD,Shoenfeld Y.Liver cystic echinococcosis and human host immune and autoimmune follow-up:A review. World J Hepatol.2017;9(30):1176-1189.[20] Jazouli M,Lightowlers M,Gauci CG,et al.Vaccination Against Hydatidosis: Molecular Cloning and Optimal Expression of the EG95NC(-) Recombinant Antigen in Escherichia coli.Protein J. 2017;36(6):472-477.[21] Alvarez Rojas CA, Gauci CG, Lightowlers MW. Antigenic differences between the EG95-related proteins from Echinococcus granulosus G1 and G6 genotypes:implications for vaccination. Parasite Immunol.2013;35(2):99-102.[22] 安梦婷,吾拉木•马木提,马海梅,等.细粒棘球蚴AgB 1、AgB2、AgB4抗原表位的特性分析[J].新疆医科大学学报, 2016,39(12): 1541-1546,1553.[23] Fu Y,Saint-André Marchal I,Marchal T,et al.Cellular immune response of lymph nodes from dogs following the intradermal injection of a recombinant antigen corresponding to a 66 kDa protein of Echinococcus granulosus.Vet Immunol Immunopathol. 2000;74(3-4):195-208.[24] Saboulard D,Lahmar S,Petavy AF,et al.The Echinococcus granulosus antigen EgA31: localization during development and immunogenic properties.Parasite Immunol.2003;25(10): 489-501.[25] Ma X,Zhou X,Zhu Y,et al.The prediction of T- and B-combined epitope and tertiary structure of the Eg95 antigen of Echinococcus granulosus.Exp Ther Med.2013;6(3):657-662.[26] Ma X,Zhao H,Zhang F,et al.Activity in mice of recombinant BCG-EgG1Y162 vaccine for Echinococcus granulosus infection.Hum Vaccin Immunother.2016;12(1):170-175.[27] Shen CM,Zhu BF,Deng YJ,et al.Allele polymorphism and haplotype diversity of HLA-A, -B and -DRB1 loci in sequence-based typing for Chinese Uyghur ethnic group. PLoS One.2010;5(11):e13458. |