Chinese Journal of Tissue Engineering Research ›› 2017, Vol. 21 ›› Issue (20): 3224-3233.doi: 10.3969/j.issn.2095-4344.2017.20.018
Previous Articles Next Articles
Molecular dynamics simulation: a new direction targeting physiological mechanisms of neuronal calcium sensor-1 protein
Zhu Yu-zhen1, Zhang Qing-wen2
- 1School of Physical Education, Shanghai Normal University, Shanghai 200234, China; 2Shanghai University of Sport, Shanghai 200438, China
-
Revised:2017-02-23Online:2017-07-18Published:2017-07-28 -
About author:Zhu Yu-zhen, M.D., School of Physical Education, Shanghai Normal University, Shanghai 200234, China -
Supported by:the Postgraduate Education Innovation Research Project of Shanghai University of Sport, No. yjscx2015003
CLC Number:
Cite this article
Zhu Yu-zhen, Zhang Qing-wen. Molecular dynamics simulation: a new direction targeting physiological mechanisms of neuronal calcium sensor-1 protein[J]. Chinese Journal of Tissue Engineering Research, 2017, 21(20): 3224-3233.
share this article
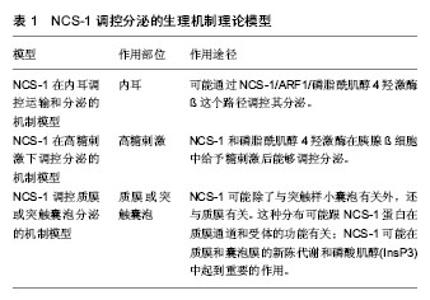
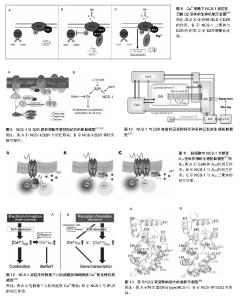
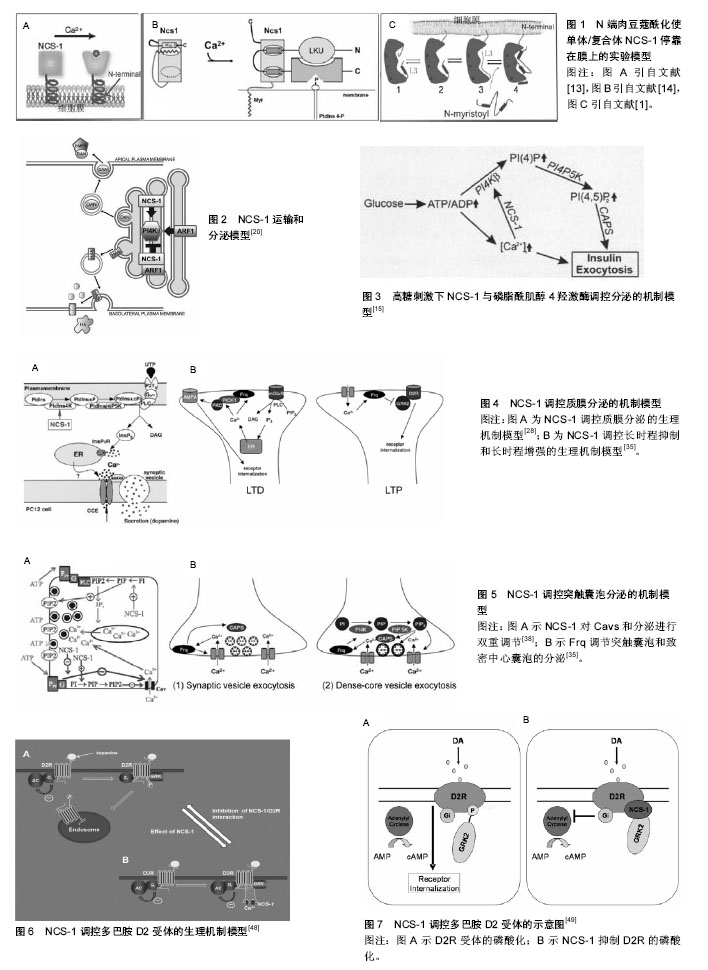
2.1 NCS-1生理机制的实验理论模型 NCS-1在不同等级生物体内分布广泛,功能表现出多样性。随着研究的深入,逐渐发现其功能发挥的部分生理机制,并提出一些实验理论模型,现将NCS-1与靶体相互作用的生理机制实验理论模型进行梳理和总结,以期进一步加深对NCS-1功能发挥的理解。 2.1.1 NCS-1停靠在膜的生理机制理论模型 NSC-1通过其N端的14碳链,主要与高尔基体膜相连,多分布于细胞核周围;少量的NCS-1因高尔基体的胞吐、分泌作用,被带到细胞膜上或胞质内的分泌囊泡膜上[2]。当失去14碳链后,NCS-1失去空间特性,弥漫整个胞质,均匀分布;胞内游离钙离子浓度升高,不影响NCS-1亚细胞的分布。在这点上,与钙传感蛋白亚家族中的海马钙素、神经钙素等截然不同[3]。 有研究指出,NCS-1在Ca2+依赖方式下上行到膜[4]。也有研究指出,N端的肉豆蔻酰化是一个脂质末端变异体,一半的脂质附属物导致了疏水性的提高,从而将蛋白质固定在膜处[5](见图1)。但通过脂质使NCS-1停靠在膜位置上的功能机制一直存在争议[6-7]。虽然NCS-1跟NCS家族其他成员一样,也具有4个EF手性位点,只有其中3个可以与Ca2+结合(见图1C中圆点),但NCS-1又与家族中的其他成员不同,其通过结合大量的配体能够与细胞内浓度发生微小变化的Ca2+进行结合和传输,并根据环境重塑其结构[8](见图1A,B);而且其可以在毫秒级时间内对Ca2+的变化作出反应,并将其迅速传导,对钙离子的结合具有“高亲和力,低容量”的特点[9]。当与Ca2+结合,并达到饱和状态时,可以和大量的结合物相互作用,这时会有一个大的构象变化,直到结构趋于稳定[10](见图1B),而构象变化又使NCS-1转变到它的活性状态,从而调节细胞内的Ca2+[11]。如果没有钙和镁,只暂时存在于活性状态中的NCS-1就不能保持一个稳定的折叠结构[12]。 NCS-1在与膜接触前,N端肉豆蔻酰呈折叠状态(见图1B,C1),并在疏水口袋内;加入Ca2+后,NCS-1构象发生改变(见图1A,B),暴露出N端肉豆蔻酰并插入膜中(图1B,C),N端甘氨酸通过N-肉豆蔻酰基转移酶的作用,使NCS-1停靠在膜的位置(见图1B,C2-4);当有小分子Pik1时,含有Ca2+的NCS-1与Pik1结合,导致Pik1形状改变,促进脂质激酶LKU与Pik1的催化部分相互作用(图1B);当缺乏配体时,蛋白C末端L3在疏水口袋内充当临时配体(见图1C3),维持蛋白结构稳定。另外,加入Ca2+后,NCS-1蛋白的N-端肉豆蔻酰促进复合体靠近膜,在膜处使NCS-1/Pik1复合体与底物(Ptdins)结合并提高磷脂酰肌醇-激酶(Ptdins 4-P)活性。 2.1.2 NCS-1调控分泌的生理机制理论模型 磷脂酰肌醇4羟激酶β能够调控酵母和哺乳动物细胞体内的从高尔基体到质膜的物质运输[15-17],也能调控PC12细胞和胰腺ß细胞的分泌[18-19]。根据前人的研究,对NCS-1与磷脂酰肌醇4羟激酶β在不同等级生物体内,不同位置调控分泌的生理机制模型进行了归纳和分类,下面分别进行阐述,见表1。"
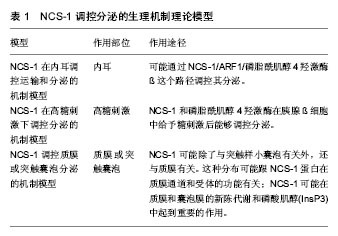
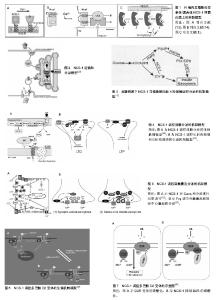
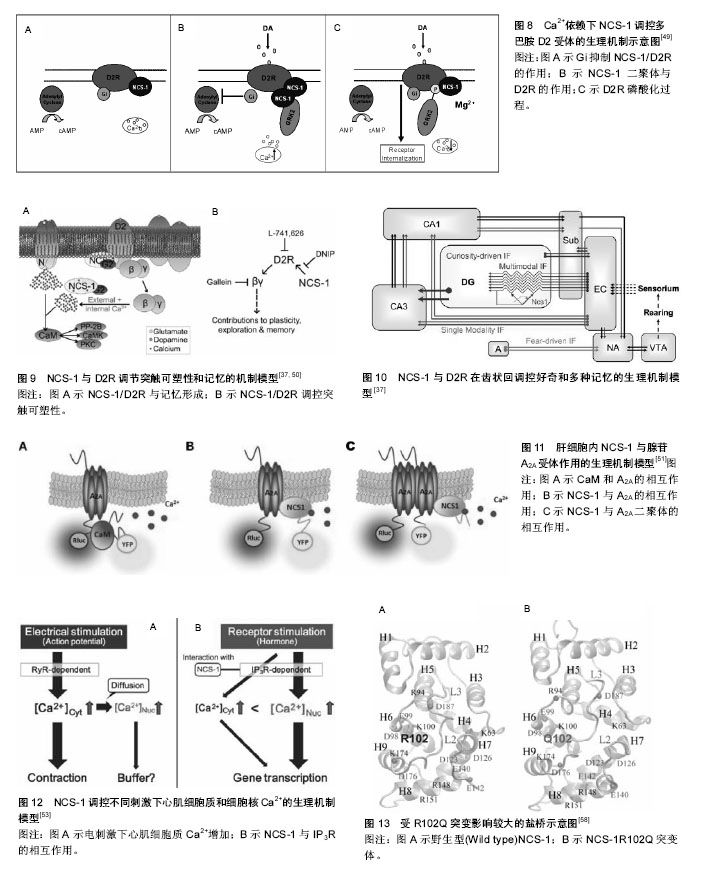
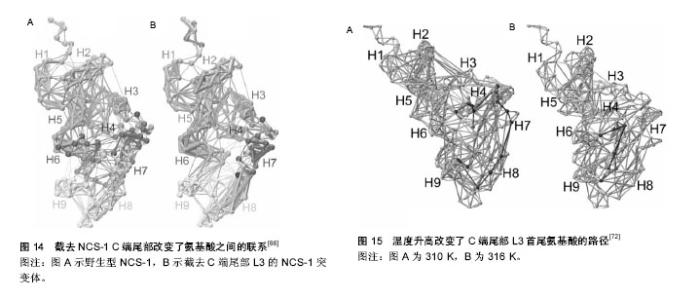
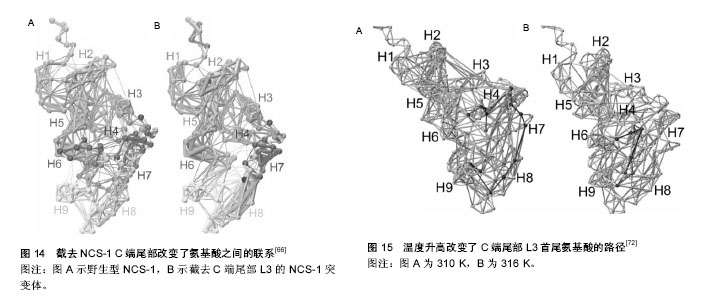
NCS-1在内耳调控运输和分泌的机制模型:Petko等[20]提出了斑马鱼内耳信号传导的途径并提出一个模型见图2[20]。NCS-1和ARF1能激活磷脂酰肌醇4羟激酶β,但NCS-1和ARF1直接相互作用能抑制磷脂酰肌醇4羟激酶β的酶活性[21-22],NCS-1和ARF1对磷脂酰肌醇4羟激酶β双向的激活/抑制可能参与了个体内耳的发育过程[20]。DAN、骨形态发生蛋白4和透明质酸是分泌分子,在半规管形成中起到重要的作用[23-27],可能通过NCS-1/ ARF1/磷脂酰肌醇4羟激酶β这个路径调控其分泌[20]。 如图2所示,在内耳中,NCS-1、ARF1和磷脂酰肌醇4羟激酶β形成了一个小的相互作用网络,这3种蛋白形成半规管调控靶蛋白分子DAN和透明质酸分泌(HAS),共分成两条途径调控分泌,一条是DAN通过顶端质膜参与骨形态发生蛋白4的分泌,另一条HAS通过基底外侧质膜调控透明质酸的分泌。 NCS-1在高糖刺激下调控分泌的机制模型:NCS-1和磷脂酰肌醇4羟激酶在胰腺ß细胞中给予糖刺激后能够调控分泌[15]。如图3所示,高糖能够提高ATP/ADP的比率,而提高的ATP/ADP会导致膜去极化,促使细胞质中的Ca2+浓度提高;而提高的ATP/ADP和Ca2+浓度能够刺激磷脂酰肌醇4羟激酶β的活性,分别通过磷脂酰肌醇4羟激酶β和磷脂酰肌醇4羟激酶 5-激酶形成PI(4)P和PI(4,5)P2,这两者能够加强Ca2+依赖激活蛋白的分泌(CAPS),从而增强胰岛素分泌。 NCS-1调控质膜或突触囊泡分泌的机制模型:NCS-1蛋白在PC12细胞中有广泛的分布,部分分布在细胞质中,部分在囊泡和管状结构中。形态学和生化结果表明,NCS-1可能除了与突触样小囊泡有关外,还与质膜有 关[28]。这种分布可能跟NCS-1蛋白在质膜通道和受体的功能有关[29],尤其是与门控Ca2+通道有关[30-32];NCS-1可能在质膜和囊泡膜的新陈代谢和磷酸肌醇(InsP3)中起到重要的作用[28]。磷脂家族在质膜中参与信号转导和囊泡形成、分泌面高尔基体网络和核内体[33]。在酵母中,NCS-1的同源体以复合体形式存在,并激活磷脂酰肌醇4羟激酶(PtdIns4K)[19],而且在真核细胞中NCS-1与PtdIns4K直接相互作用[34]。在脊椎动物的PC12细胞中,NCS-1在不影响PtdIns的情况下,明显提高了PtdIns(4)P 和PtdIns(4,5)P2的水平,促进Ins(1,4,5)P3的增多,从而与G蛋白偶联受体(GPCRs)的兴奋剂UTP进行反应。NCS-1可能促进质膜或其他细胞器中PtdIns4K的激活和更新,提高PtdIns的新陈代谢和合成,增加信号分子肌醇1,4,5,-三磷酸(Ins(1,4,5)P3),通过肌醇1,4,5,-三磷酸受体(InsP3R)增加影响细胞内Ca2+信号和胞外分泌。 图4A所示,NCS-1结合到位于质膜或突触囊泡的PtdIns4K,并增加其活性,这就提高PtdIns(4)P和PtdIns(4,5)P2(PIP2)在局部位置的水平,其充当了磷脂酶C的底物,导致肌醇三磷酸Ins(1,4,5)P3(IP3)和二脂酰甘油的增加;通过Ins(1,4,5)P3受体和CCE,增加的Ins(1,4,5)P3(IP3)会诱发Ca2+的反应,从而加强分泌反应。但NCS-1和PtdIns4K相互作用的动力学机制尚不完全清楚[28],需要从更微观的角度深入研究。 图4B对哺乳动物的中枢神经系统突触后的Frq调节长时程抑制和长时程增强的机制进行了研究[35],并提出模型。图4B左图中,当亲代谢性谷氨酸盐受体(mGluR)激活后,4,5-二磷酸磷脂酰肌醇(PIP2)充当了磷脂酶C的底物,引起肌醇三磷酸Ins(1,4,5)P3(IP3)和二脂酰甘油的增加,导致内质网(ER)释放Ca2+。Frq和蛋白激酶C相互作用蛋白1(PICK1)与从内部钙离子储存器ER中释放出来的Ca2+结合,Frq可能与PICK1形成复合物和蛋白激酶(PKC),这就引起α-氨基-3-羟基-5-甲基-4-异恶唑丙酸受体(AMPAR)的磷酸化和内化,导致长时程抑制[36];图4B右图中,Frq结合Ca2+后,结合到G偶联蛋白受体激酶2(GRK2),抑制多巴胺D2受体(D2R)的磷酸化,提高D2R的表面表达,导致长时程增强[37]。 另外,NCS-1作为神经传递的中介,对Cavs和分泌进行双重调节。图5A中,实线描述了NCS-1对分泌和Cavs通道的影响。其中,带加号的箭头表明活性提高;带减号的箭头表明活性降低。NCS-1通过磷脂酰肌醇4羟激酶Ⅲß的活性和PIP2水平的提高能够加强分泌,导致调节内部Ca2+储备释放的IP3提高。Ca2+水平的提高将会提高囊泡的运动,从而促使囊泡与质膜融合;另外,质膜中的PIP2本身也能够促进囊泡的融合和运动,PIP2还具有稳定Cavs通道(N-type)的功能,并通过NCS-1提高磷脂酰肌醇4羟激酶Ⅲß的活性和PIP2水平而加强这一功能。NCS-1能够通过嘌呤GPCR抑制通路的信号,从而抑制Cavs(P/Q-type)。数据表明,PTX敏感的G蛋白和Src样激酶也参与到NCS-1抑制P/Q-type Cavs过程中,但NCS-1抑制P/Q-type Cavs的具体机制还不清楚[38]。 图5B显示了Frq调节突触囊泡和致密中心囊泡分泌的机制模型。图5B(1)中,Frq结合Ca2+直接或者通过Ca2+依赖激活蛋白(CAPS)调控Ca2+信号,从而调控突触囊泡的分泌[31-32,39]。图5B(2)中Frq结合Ca2+并激活磷脂酰肌醇4羟激酶ß,从而加强磷脂酰肌醇4磷酸盐(PIP)和磷脂酰肌醇4,5二磷酸(PIP2)水平[4,21-22,28,40-44]。磷酸肌醇(PIP和PIP2)水平提高,能够促进致密中心囊泡的分泌。 2.1.3 NCS-1调控多巴胺D2受体的生理机制模型 多巴胺是哺乳动物脑内其中一种重要的神经递质。研究显示,多巴胺释放引起的神经反应在很多生理功能中起到作用,包括动作控制、情绪、认知、神经内分泌的控制、参与奖赏活动等。为了执行对多巴胺释放的各种反应,脑内有4条主要的多巴胺能通路:中脑缘通路,中脑皮质通路,黑质纹状体通路,结状漏斗形通路[45]。由多巴胺神经递质产生的多种功能不仅依赖于4条主要的神经通路,而且在这些通路内,不同的信号由不同的多巴胺受体传输。到目前为止,5个已知的多巴胺受体(DR)亚型有D1、D2、D3、D4和D5。5个受体亚型是7次横跨细胞膜的G蛋白偶联受体(GPCRs),分为两个多巴胺受体家族,D1家族样受体包括D1和D5两个亚型,D2家族样受体包括D2、D3及D4[46]。其中多巴胺D2受体包含415个氨基酸,由8个外显子与7个内含子组成,属于G蛋白偶联受体家族,其肽链7次跨越细胞膜,N端在胞外,C端在胞内。多巴胺D2受体通过细胞质内C端尾部16个氨基酸与NCS-1结合,可能影响多巴胺D2受体内化,并控制多巴胺D2受体表面表达[47]。如图6A所示,多巴胺通过D2R发挥作用,受到G蛋白Gi的抑制从而引起腺苷酸环化酶的抑制;D2R通过G蛋白偶联受体激酶进行磷酸化,磷酸化了的D2R在核内体里(endosome)通过细胞的内吞作用而内化,进行脱敏。在这一过程中,NCS-1的作用见图6B,含有Ca2+的NCS-1通过与D2R受体C端尾部和G蛋白偶联受体激酶结合,抑制D2R受体的磷酸化,导致D2R非但没有内在化反而保持在细胞表面;而NCS-1/D2R相互作用的抑制允许D2R受体脱敏[48]。 图7更清晰地显示出NCS-1与D2R受体相互作用的生理机制。多巴胺释放通过D2R受体发挥作用,D2R通过G蛋白偶联受体激酶2进行磷酸化,从而导致D2R内在化,刺激腺苷酸环化酶的活性,使基因表达调控物质AMP不断生成腺苷-3’,5’-环化一磷酸cAMP(见图7A);当NCS-1通过G蛋白偶联受体激酶2抑制D2R的磷酸化,D2R继续传导信号,但抑制腺苷酸环化酶的活性(见图7B)。 当D2R受到G蛋白Gi的抑制引起腺苷酸环化酶抑制的情况下,即使NCS-1单体结合到D2R上,Ca2+储存器也不受D2R活动的影响(见图8A);当D2R通过Gi抑制腺苷酸环化酶的活性时,内质网的Ca2+储存器释放Ca2+,引起细胞内Ca2+水平提高会促进NCS-1二聚体结合到D2R上(见图8B);当细胞内Ca2+水平下降到NCS-1二聚体形成的阈值以下,G蛋白偶联受体激酶2使D2R磷酸化,从而使激活的D2R内在化(见图8C)。 NCS-1与多巴胺D2受体相互作用调节突触可塑性和记忆的机制模型见图9。NCS-1通过抑制G蛋白偶联受体激酶G蛋白偶联受体激酶2(G2),导致D2R磷酸化,提高了D2R在膜表面的表达,阻止D2R内在化。在电刺激或者行为刺激下,D2R导致更高的Ca2+反应,可能是提高记忆形成的机制[50](见图9A)。NCS-1调控D2R表面表达,这一过程受到D2R/NCS-1相互作用肽DNIP的阻碍;D2R的活性是突触可塑性加强所必需的,这一过程受到L-741-626的阻碍;G蛋白ß和Y信号也能加强突触可塑性,这一过程受到焦口酚酞(Gallein)的阻碍(见图9B)。 NCS-1与D2R在齿状回调控好奇和多种记忆的生理机制模型[37],见图10。来自感觉中枢(sensorium)的多种信息都在内嗅皮质(EC)加以整合和投射到齿状回(DG)中,NCS-1降低了LTP的感应阈值,加强了内嗅皮质-齿状回的LTP,促进快速获取和空间记忆;单形式信息可能通过內嗅皮质直接投射到海马(CA)CA1和CA3中;负责探索和好奇行为在齿状回出现,以及通过內嗅皮质和/或海马三突触回路(CA3-CA1-Sub)投射到伏隔核(NA);在NA和腹侧被盖区(VTA)之间的双向信号传送导致饲养行为(rearing)。而饲养行为反过来会导致更丰富的感觉,这可能是一个正反馈通路;由恐惧驱使的饲养行为在杏仁核(A)中出现,在图10模型中这一行为未与其他脑区发生联系。 2.1.4 肝细胞内NCS-1与腺苷A2A受体作用的生理机制模型 腺苷A2A受体可以与钙调蛋白和NCS-1相互作 用[51]。发光供体氧化底质腔肠素H发出荧光,当供体和受体之间的距离约为4.4 nm时,其可以转为荧光受体(YFP)[52]。图11中比较了人胚肾HEK193T细胞共表达A2A融合到海肾萤光素酶(Rluc)和CaM中,NCS-1融合到YFP中(A-C)。当CaM-YFP表达增加时,CaM和A2A有特定的相互作用(见图11A);当A2A-Rluc数量一定,NCS-1-YFP共表达数量的增加时,NCS-1与A2A有特定的相互作用(见图11B);在Ca2+依赖模式下,NCS-1与A2A二聚体相互作用,并调控腺苷A2A受体的信号(见图11C)。 2.1.5 NCS-1调控不同刺激下心肌细胞质和细胞核Ca2+的生理机制模型 Nakao等[53]对NCS-1调控不同刺激下心肌细胞质和细胞核Ca2+的生理机制模型进行了推测(见图12)。在电刺激下,通过Ca2+诱导的Ca2+释放RyR依赖机制,细胞质的Ca2+从肌质网(SR)释放出来,细胞质Ca2+([Ca2+]Cyt)不断扩散到细胞核中,细胞核Ca2+([Ca2+]Nuc)增加。增加的细胞质Ca2+导致肌肉收缩,细胞核可能不断缓冲细胞质内不断增加的Ca2+(见图12A);另外,受体刺激可能通过IP3R与NCS-1相互作用,优先增加细胞核Ca2+;受体刺激诱导增加细胞质内的Ca2+可能加强基因转录,导致心肌肥大(见图12B)。而NCS-1在受体刺激调控细胞质内的Ca2+中与IP3R相互作用而起到重要的作用。 根据实验研究结果提出蛋白可能的生理机制模型,对深入理解蛋白功能的发挥有很好的启发作用和参考价值,但在现有实验条件下,这些生理机制模型虽已提出,但尚无法进行更为深入的研究,更无从得以验证和确认,导致研究暂时无法继续向纵深方向推进。 2.2 NCS-1生理机制的结构特征 结构决定功能,NCS-1生理功能的正常发挥依赖于结构的完整性和稳定性。对蛋白结构变化引起生理功能改变的观察,实验上多利用荧光技术、核磁共振和化学位移等方法和手段,得出一定的结果和结论,而且通过分子动力学模拟的方法,对实验结果和结论作进一步的深入研究和探讨,不仅对目前存在的矛盾和争论有一定的解释,而且还给出了蛋白结构中更加微观的机制变化,为实验研究提供新的视角和启示。 2.2.1 NCS-1R102Q突变导致结构的变化引起生理功能改变 在自闭症患者基因测序中识别出组成NCS-1 190个氨基酸中,第102个氨基酸精氨酸ARG102突变成谷氨酰胺Glu102(R102Q)[54]。R102Q突变对蛋白结构既有远距离的影响,也有局部的改变,尤其是螺旋H6、H9和C端尾部L3,但R102Q突变引起结构改变有两种冲突的机制解释。一项核磁共振实验研究表明,R102Q突变影响NCS-1蛋白的结构,尤其提高了C端尾部构象交换的范围[55];但另一项高分辨率核磁共振结构和化学位移测量研究表明,R102Q突变降低了C端尾部L3的活 动[1]。对R102Q突变和C端尾部活动的描述通常能够反映蛋白的基本特性[56-57]。为从原子-分子层面,对NCS-1R102Q突变导致蛋白结构的变化有更微观的解释,有研究采用分子动力学的方法,发现R102Q突变导致某些盐桥的改变[8,58-59](见图13),对蛋白结构柔性的改变和L3在疏水口袋中的位置起到重要的作用,该研究结果与实验上观察到的R102Q突变降低了C端尾部L3的活动结果一致[1],可能会阻碍人类NCS-1蛋白与其配体的结合,影响其功能的发挥;并从蛋白结构的视角,为与R102Q突变有关的自闭症疾病提供原子水平的解释,为实验研究提供参考和启示。 2.2.2 C端尾部对NCS-1蛋白生理功能发挥的影响NCS-1通过疏水口袋中的结合位点与配体结合,发挥其功能。在NCS-1复合体中,疏水口袋中的结合位点被Pik1片段和聚乙二醇分子占据[10,60-62],对蛋白结构起到稳定作用[1];当缺乏配体的时候,C端尾部L3充当配体直接与疏水口袋中的位点结合,维持蛋白结构稳定[1];当配体到达疏水口袋时,L3会让出疏水口袋,使其与配体结合[1,8,63]。NCS-1能够调节多巴胺D2/D3受体[64],D2受体的C端能结合到NCS-1的疏水口袋中[47,65],而且NCS-1的C端尾部对D2受体的结合具有重要的作用[47]。实验结果显示,去掉L3会导致NCS-1整体结构不稳定[1],但在原子层面上,C端尾部对NCS-1构象影响的详细机制还不清楚。作者采用分子动力学模拟的方法,首次对人类野生型和截去C端尾部L3突变体NCS-1在水溶液的结构特征进行分析,结果显示,截去L3,NCS-1蛋白保持了二级结构,但提高了C段和远端N段的结构柔性,验证了实验结果;并且观察到截去L3削弱了N段和C段氨基酸之间的联系(见图14);破坏了蛋白盐桥网络,导致蛋白整个结构和疏水口袋同时膨胀[66]。分子动力学模拟从微观的视角表明,L3对NCS-1构象动力学起到重要的作用,截去L3引起结构的变化可能会影响与配体的相互作用,进而影响其生理功能的发挥。 2.2.3 温度升高影响NCS-1生理功能发挥 NCS-1是调控动物对温度变化做出相应反应的重要因素之一。温度对人类机体影响的分子机制还不完全清楚,但国内外学者对线虫的趋温性做了较多的研究[67]。某些温度感受器对微小的温度波动(<0.01 ℃)很敏感,这些感受器通过适应,对大幅度室温变化保持一定的敏感性。在AFD神经元中,鸟苷酸环化酶受体(GCY-8)、磷酸二酯酶(PDE-2)和NCS-1调控线虫的趋温性[68]。在径向温度梯度中,线虫沿着等温线,朝培养温度移动,线虫的这种学习和记忆是NCS-1通过Ca2+信号调整的[69-70]。最近,调控线虫温度依赖性运动的关键因素被识别出来。研究表明,线虫运动的改变是由NCS-1引起的,尤其是完全暴露于水的疏水口袋的完整性起到重要的作用[71],但其详细细节和微观机制尚不清楚。线虫和人类NCS-1的氨基酸序列相似性为75.4%,但温度改变是否影响人类NCS-1的结构尚不清楚。假设温度升高会影响人蛋白C端尾部L3在疏水口袋的位置,采用分子动力学模拟的方法,比较人类NCS-1在正常体温和极限体温下的构象变化,确定温度变化是否影响C端尾部L3在疏水口袋中的位置[72]。结果表明,人类NCS-1对溶液温度非常敏感。温度升高,L3从蜷缩状态转为伸展状态,更多地占据疏水口袋,可能会遮挡住疏水口袋内的结合位点,阻碍其与配体结合,影响蛋白功能的正常发挥。其中,氨基酸之间的联系及其路径改变是引起蛋白构象变化的主要原因(见图15)。从原子-分子层面,一方面验证了实验上观察到的结果,另一方面从更微观的视角提出实验上观察到现象的关键性因素,深刻揭示了温度变化对人类NCS-1关键结构的影响,为实验研究提供新的视角和启示。"
| [1] Heidarsson PO, Bjerrum-Bohr IJ, Jensen GA, et al. The C-terminal tail of human neuronal calcium sensor 1 regulates the conformational stability of the Ca 2+- activated state.J Mol Biol. 2012;417: 51-64.[2] Hilfiker S. Neuronal calcium sensor-1: a multifunctional regulator of secretion.Biochem Soc Trans. 2003;31: 828-832.[3] O'Callaghan DW, Ivings L, Weiss JL, et al. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca2+ signal transduction.J Biol Chem. 2002;277: 14227-14237.[4] McFerran BW, Weiss JL, Burgoyne RD. Neuronal Ca2+ sensor 1. Characterization of the myristoylated protein, its cellular effects in permeabilized adrenal chromaffin cells, Ca2+-independent membrane association, and interaction with binding proteins, suggesting a role in rapid Ca2+ signal transduction.J Biol Chem. 1999;274: 30258-30265.[5] Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins.Biochemistry. 1993;32: 10436-10443.[6] O'Callaghan DW, Burgoyne RD. Identification of residues that determine the absence of a Ca2+/myristoyl switch in neuronal calcium sensor-1.J Biol Chem. 2004;279: 14347-14354.[7] Strahl T, Grafelmann B, Dannenberg J,et al. Conservation of regulatory function in calcium-binding proteins: human frequenin (neuronal calcium sensor-1) associates productively with yeast phosphatidylinositol 4-kinase isoform, Pik1.J Biol Chem. 2003;278: 49589-49599.[8] Bellucci L, Corni S, Di Felice R, et al. The Structure of Neuronal Calcium Sensor-1 in SolutionRevealed by Molecular Dynamics Simulations. PLoS One. 2013;8: e74383.[9] 张占军,李澎,邱蕾,等. 神经元钙传感蛋白的研究进展及其在脑缺血中的作用[J].中国药理学通报. 2009;25(1): 12-15.[10] Ames B, Hendricks KB, Strahl T, et al. Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae.Biochemistry. 2000;39: 12149-12161.[11] Ikura M. Calcium binding and conformational response in EF-hand proteins.Trends Biochem Sci. 1996;21: 14-17.[12] Aravind P, Chandra K, Reddy PP, et al. Regulatory and structural EF-hand motifs of neuronal calcium sensor-1: Mg2+ modulates Ca2+ binding, Ca2+-induced conformational changes, and equilibrium unfolding transitions.J Mol Biol. 2008;376:1100-1115.[13] Haynes LP, Burgoyne RD. Unexpected tails of a Ca2+ sensor. Nat Chem Biol. 2008;4: 90-91.[14] Lim S, Strahl T, Thorner J, et al. Structure of a Ca2+-myristoyl switch protein that controls activation of a phosphatidylinositol 4-kinase in fission yeast.J Biol Chem. 2011;286: 12565- 12577.[15] Gromada J, Bark C, Smidt K, et al. Neuronal calcium sensor-1 potentiates glucose-dependent exocytosis in pancreatic beta cells through activation of phosphatidylinositol 4-kinase beta.Proc Natl Acad Sci U S A. 2005;102:10303-10308.[16] Bruns JR, Ellis MA, Jeromin A, et al. Multiple roles for phosphatidylinositol 4-kinase in biosynthetic transport in polarized Madin-Darby canine kidney cells.J Biol Chem. 2002;277: 2012-2018.[17] Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi.Nat Cell Biol. 1999;1: 523-525.[18] Weisz OA, Gibson GA, Leung SM, et al. Overexpression of frequenin, a modulator of phosphatidylinositol 4-kinase, inhibits biosynthetic delivery of an apical protein in polarized madin-darby canine kidney cells.J Biol Chem. 2000;275: 24341-24347.[19] Hendricks KB, Wang BQ, Schnieders EA, et al. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase.Nat. Cell. Biol. 1999;1: 234-241.[20] Petko JA, Kabbani N, Frey C, et al. Proteomic and functional analysis of NCS-1 binding proteins reveals novel signaling pathways required for inner ear development in zebrafish. BMC neuroscience. 2009;10: 27.[21] Haynes LP, Sherwood MW, Dolman NJ, et al. Specificity, promiscuity and localization of ARF protein interactions with NCS-1 and phosphatidylinositol-4 kinase-III beta.Traffic. 2007;8: 1080-1092.[22] Haynes LP, Thomas GM, Burgoyne RD. Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase beta and trans-Golgi network-plasma membrane traffic.J Biol Chem. 2005;280: 6047-6054.[23] Chang W, Lin Z, Kulessa H, et al. Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements.PLoS Genet. 2008;4: e1000050.[24] Yamanishi T, Katsu K, Funahashi J, et al. Dan is required for normal morphogenesis and patterning in the developing chick inner ear. Dev Growth Differ. 2007; 49(1):13-26.[25] Busch-Nentwich E, Sollner C, Roehl H, et al. The deafness gene dfna5 is crucial for ugdh expression and HA production in the developing ear in zebrafish.Development. 2004;131: 943-951.[26] Haddon CM, Lewis JH. Hyaluronan as a propellant for epithelial movement: the development of semicircular canals in the inner ear of Xenopus.Development. 1991;112: 541-550.[27] Omata Y, Nojima Y, Nakayama S, et al. Role of Bone morphogenetic protein 4 in zebrafish semicircular canal development.Dev Growth Differ. 2007;49: 711-719.[28] Koizumi S, Rosa P, Willars GB, et al. Mechanisms underlying the neuronal calcium sensor-1-evoked enhancement of exocytosis in PC12 cells.J Biol Chem. 2002;277: 30315- 30324.[29] Nef S, Fiumelli H, de Castro E,et al. Identification of neuronal calcium sensor (NCS-1) possibly involved in the regulation of receptor phosphorylation.J Recept Signal Transduct Res. 1995;15: 365-378.[30] Weiss JL, Archer DA, Burgoyne RD. Neuronal Ca2+ sensor-1/frequenin functions in an autocrine pathway regulating Ca2+ channels in bovine adrenal chromaffin cells.J Biol Chem. 2000;275: 40082-40087.[31] Wang CY, Yang F, He X, et al. Ca2+ binding protein frequenin mediates GDNF-induced potentiation of Ca2+ channels and transmitter release.Neuron. 2001;32: 99-112.[32] Tsujimoto T, Jeromin A, Saitoh N, et al. Neuronal calcium sensor 1 and activity-dependent facilitation of P/Q-type calcium currents at presynaptic nerve terminals.Science. 2002;295: 2276-2279.[33] De Camilli P, Emr SD, McPherson PS,et al. Phosphoinositides as regulators in membrane traffic.Science. 1996;271:1533-1539.[34] Pan CY, Jeromin A, Lundstrom K, et al. Alterations in exocytosis induced by neuronal Ca2+ sensor-1 in bovine chromaffin cells.J Neurosci. 2002;22: 2427-2433.[35] Dason JS, Romero-Pozuelo J, Atwood HL, et al. Multiple roles for frequenin/NCS-1 in synaptic function and development. Mol Neurobiol. 2012;45: 388-402.[36] Jo J, Heon S, Kim MJ, et al. Metabotropic glutamate receptor-mediated LTD involves two interacting Ca2+ sensors, NCS-1 and PICK1.Neuron. 2008;60: 1095-1111.[37] Saab BJ, Georgiou J, Nath A, et al. NCS-1 in the dentate gyrus promotes exploration, synaptic plasticity, and rapid acquisition of spatial memory.Neuron. 2009;63: 643-656.[38] Weiss JL, Hui H, Burgoyne RD. Neuronal calcium sensor-1 regulation of calcium channels, secretion, and neuronal outgrowth.Cell Mol Neurobiol. 2010;30:1283-1292.[39] Dason JS, Romero-Pozuelo J, Marin L, et al. Frequenin/NCS-1 and the Ca2+-channel alpha1-subunit co-regulate synaptic transmission and nerve-terminal growth.J Cell Sci. 2009;122:4109-4121.[40] de Barry J, Janoshazi A, Dupont JL, et al. Functional implication of neuronal calcium sensor-1 and phosphoinositol 4-kinase-beta interaction in regulated exocytosis of PC12 cells.J Biol Chem. 2006;281: 18098-18111.[41] McFerran BW, Graham ME, Burgoyne RD. Neuronal Ca2+ sensor 1, the mammalian homologue of frequenin, is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules.J Biol Chem. 1998; 273: 22768-22772.[42] Taverna E, Francolini M, Jeromin A, et al. Neuronal calcium sensor 1 and phosphatidylinositol 4-OH kinase beta interact in neuronal cells and are translocated to membranes during nucleotide-evoked exocytosis.J Cell Sci. 2002;115: 3909-3922.[43] Scalettar BA, Rosa P, Taverna E,et al. Neuronal calcium sensor-1 binds to regulated secretory organelles and functions in basal and stimulated exocytosis in PC12 cells.J Cell Sci. 2002;115: 2399-2412.[44] Rajebhosale M, Greenwood S, Vidugiriene J, et al. Phosphatidylinositol 4-OH kinase is a downstream target of neuronal calcium sensor-1 in enhancing exocytosis in neuroendocrine cells.J Biol Chem. 2003;278: 6075-6084.[45] Missale C, Nash SR, Robinson SW, et al. Dopamine receptors: from structure to function.Phys Rev. 1998;78: 189-225.[46] Kebabian JW, Calne DB. Multiple receptors for dopamine. Nature. 1979;277: 93-96.[47] Lian LY, Pandalaneni SR, Patel P, et al. Characterisation of the interaction of the C-terminus of the dopamine D2 receptor with neuronal calcium sensor-1.PLoS One. 2011;6: e27779.[48] Burgoyne RD, Haynes LP. Sense and specificity in neuronal calcium signalling.Biochim Bbiophys Acta. 2015;1853: 1921-1932.[49] Woll MP. Chareacterization of the D2 dopamine receptor/neuronal calcium sensor-1 interaction, In Department of Pharmacology, The Pennsylvania State University, 2009.[50] Saab BJ. The synaptic role of neuronal calcium seneor 1 in dentate gyrus plasticity, curiosity and spatial memory, In Department of molecular genetics, University of Toronto, 2010.[51] Navarro G, Hradsky J, Lluis C, et al. NCS-1 associates with adenosine A(2A) receptors and modulates receptor function. Front Mol Neurosci. 2012;5: 53.[52] Dacres H, Wang J, Dumancic MM, et al. Experimental determination of the Forster distance for two commonly used bioluminescent resonance energy transfer pairs. Anal Chem. 2010;82(1):432-435. [53] Nakao S, Wakabayashi S, Nakamura TY. Stimulus-dependent regulation of nuclear Ca2+ signaling in cardiomyocytes: a role of neuronal calcium sensor-1.PLoS One. 2015;10: e0125050.[54] Piton A, Michaud JL, Peng H, et al. Mutations in the calcium-related gene IL1RAPL1 are associated with autism.Hum Mol Genet. 2008;17: 3965-3974.[55] Handley MT, Lian LY, Haynes LP, et al. Structural and functional deficits in a neuronal calcium sensor-1 mutant identified in a case of autistic spectrum disorder.PLoS One. 2010;5: e10534.[56] Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins?Proteins. 2004;57: 433-443.[57] Ma B, Tsai CJ, Haliloglu T, et al. Dynamic allostery: linkers are not merely flexible.Structure. 2011;19: 907-917.[58] Zhu Y, Wu Y, Luo Y, et al. R102Q mutation shifts the salt-bridge network and reduces the structural flexibility of human neuronal calcium sensor-1 protein.J Phys Chem B. 2014;118: 13112-13122.[59] 朱玉珍,张庆文. 神经元钙传感蛋白R102Q突变体的动力学构象特征[J].中国组织工程研究,2015,19(2): 225-230.[60] Lim S, Strahl T, Thorner J, et al. Structure of a Ca2+-myristoyl switch protein that controls activation of a phosphatidylinositol 4-kinase in fission yeast. J Biol Chem. 2011;286: 12565- 12577.[61] Strahl T, Huttner IG, Lusin JD, et al. Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1). J Biol Chem. 2007;282: 30949-30959.[62] Bourne Y, Dannenberg J, Pollmann V,et al. Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1).The Journal of biological chemistry. 2001;276: 11949-11955.[63] Ames JB, Lim S. Molecular structure and target recognition of neuronal calcium sensor proteins.Biochim Biophys Acta. 2012; 1820: 1205-1213.[64] Kabbani N, Negyessy L, Lin R, et al. Interaction with neuronal calcium sensor NCS-1 mediates desensitization of the D2 dopamine receptor.J. Neurosci. 2002;22: 8476-8486.[65] Woll MP, De Cotiis DA, Bewley MC, et al. Interaction between the D2 dopamine receptor and neuronal calcium sensor-1 analyzed by fluorescence anisotropy.Biochemistry. 2011;50: 8780-8791.[66] Zhu Y, Yang S, Qi R, et al. Effects of the C-terminal tail on the conformational dynamics of human neuronal calcium sensor-1 protein.J Phys Chem. 2015;119: 14236-14244.[67] Kimata T, Sasakura H, Ohnishi N, et al. Thermotaxis of C. elegans as a model for temperature perception, neural information processing and neural plasticity.Worm. 2012;1: 31-41.[68] Wang D, O'Halloran D, Goodman MB. GCY-8, PDE-2, and NCS-1 are critical elements of the cGMP-dependent thermotransduction cascade in the AFD neurons responsible for C. elegans thermotaxis. J Gener Physiol. 2013;142: 437-449.[69] Gomez M, De Castro E, Guarin E, et al. Ca2+ signaling via the neuronal calcium sensor-1 regulates associative learning and memory in C. elegans. Neuron. 2001;30: 241-248.[70] Ye HY, Ye BP, Wang DY. Evaluation of the long-term memory for thermosensation regulated by neuronal calcium sensor-1 in Caenorhabditis elegans.Neurosci Bull. 2008;24: 1-6.[71] Martin VM, Johnson JR, Haynes LP, et al. Identification of key structural elements for neuronal calcium sensor-1 function in the regulation of the temperature-dependency of locomotion in C. elegans. Mol Brain. 2013;6: 39.[72] Zhu Y, Ma B, Qi R, et al. Temperature-dependent conformational properties of human neuronal calcium sensor-1 protein revealed by all-atom simulations.J Phys Chem B. 2016;120: 3551-3559. |
| [1] | Yao Xiaoling, Peng Jiancheng, Xu Yuerong, Yang Zhidong, Zhang Shuncong. Variable-angle zero-notch anterior interbody fusion system in the treatment of cervical spondylotic myelopathy: 30-month follow-up [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1377-1382. |
| [2] | An Weizheng, He Xiao, Ren Shuai, Liu Jianyu. Potential of muscle-derived stem cells in peripheral nerve regeneration [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1130-1136. |
| [3] | Zhang Jinglin, Leng Min, Zhu Boheng, Wang Hong. Mechanism and application of stem cell-derived exosomes in promoting diabetic wound healing [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 1113-1118. |
| [4] | He Yunying, Li Lingjie, Zhang Shuqi, Li Yuzhou, Yang Sheng, Ji Ping. Method of constructing cell spheroids based on agarose and polyacrylic molds [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 553-559. |
| [5] | He Guanyu, Xu Baoshan, Du Lilong, Zhang Tongxing, Huo Zhenxin, Shen Li. Biomimetic orientated microchannel annulus fibrosus scaffold constructed by silk fibroin [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 560-566. |
| [6] | Chen Xiaoxu, Luo Yaxin, Bi Haoran, Yang Kun. Preparation and application of acellular scaffold in tissue engineering and regenerative medicine [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 591-596. |
| [7] | Kang Kunlong, Wang Xintao. Research hotspot of biological scaffold materials promoting osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 597-603. |
| [8] | Shen Jiahua, Fu Yong. Application of graphene-based nanomaterials in stem cells [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 604-609. |
| [9] | Zhang Tong, Cai Jinchi, Yuan Zhifa, Zhao Haiyan, Han Xingwen, Wang Wenji. Hyaluronic acid-based composite hydrogel in cartilage injury caused by osteoarthritis: application and mechanism [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 617-625. |
| [10] | Li Hui, Chen Lianglong. Application and characteristics of bone graft materials in the treatment of spinal tuberculosis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 626-630. |
| [11] | Gao Cangjian, Yang Zhen, Liu Shuyun, Li Hao, Fu Liwei, Zhao Tianyuan, Chen Wei, Liao Zhiyao, Li Pinxue, Sui Xiang, Guo Quanyi. Electrospinning for rotator cuff repair [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 637-642. |
| [12] | Guan Jian, Jia Yanfei, Zhang Baoxin , Zhao Guozhong. Application of 4D bioprinting in tissue engineering [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(3): 446-455. |
| [13] | Liu Jiali, Suo Hairui, Yang Han, Wang Ling, Xu Mingen. Influence of lay-down angles on mechanical properties of three-dimensional printed polycaprolactone scaffolds [J]. Chinese Journal of Tissue Engineering Research, 2022, 10(16): 2612-2617. |
| [14] | Huang Bo, Chen Mingxue, Peng Liqing, Luo Xujiang, Li Huo, Wang Hao, Tian Qinyu, Lu Xiaobo, Liu Shuyun, Guo Quanyi . Fabrication and biocompatibility of injectable gelatin-methacryloyl/cartilage-derived matrix particles composite hydrogel scaffold [J]. Chinese Journal of Tissue Engineering Research, 2022, 10(16): 2600-2606. |
| [15] | Fang Xiaoyang, Tang Tian, Wang Nan, Qian Yuzhang, Xie Lin. Repair and regenerative therapies of the annulus fibrosus [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(10): 1582-1587. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||