Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (22): 5728-5738.doi: 10.12307/2026.175
Previous Articles Next Articles
Establishing a diagnostic model for recurrent spontaneous abortion based on the levels of autophagy-related genes in the endometrium
Tang Cen, Hu Wanqin
- Department of Obstetrics, Kunming Medical University Second Affiliated Hospital, Kunming 650101, Yunnan Province, China
-
Received:2025-04-03Accepted:2025-09-11Online:2026-08-08Published:2025-12-26 -
Contact:Hu Wanqin, Chief physician, Department of Obstetrics, Kunming Medical University Second Affiliated Hospital, Kunming 650101, Yunnan Province, China -
About author:Tang Cen, MS candidate, Department of Obstetrics, Kunming Medical University Second Affiliated Hospital, Kunming 650101, Yunnan Province, China -
Supported by:the National Natural Science Foundation of China, No. 82060294 (to HWQ)
CLC Number:
Cite this article
Tang Cen, Hu Wanqin. Establishing a diagnostic model for recurrent spontaneous abortion based on the levels of autophagy-related genes in the endometrium[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(22): 5728-5738.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
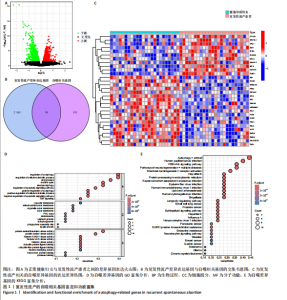
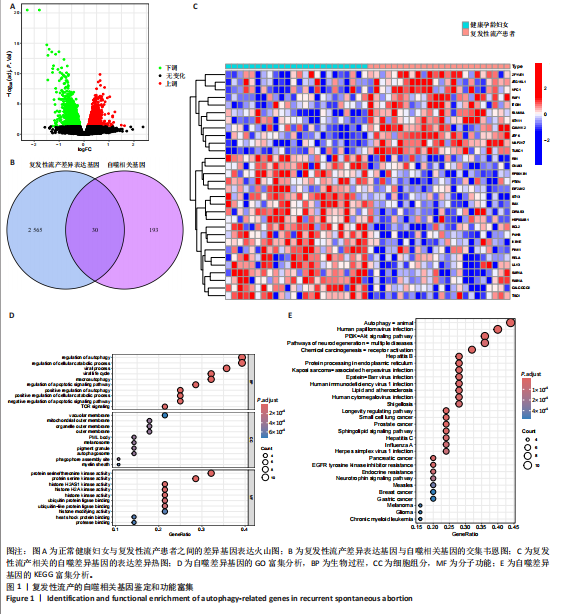
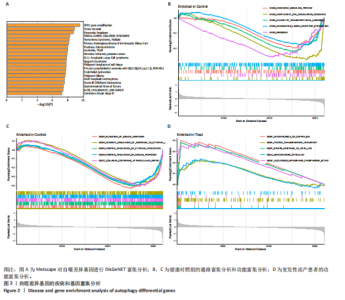
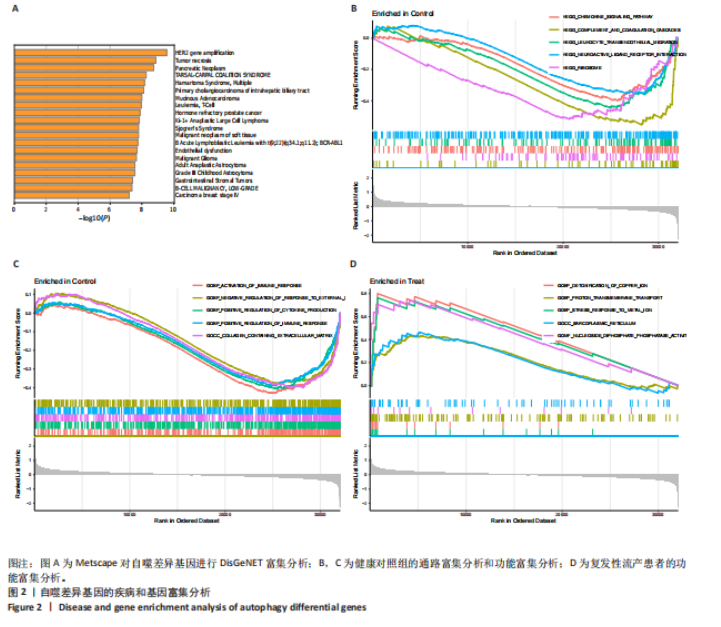
2.1 复发性流产的自噬相关基因的鉴定和功能富集 对数据集GSE165004样本数据进行差异表达分析,共筛选出2 565个差异表达基因,具体可见火山图(图1A)。将这些样本差异表达基因与HADb数据库收集的193个自噬相关基因进行比较,鉴定出30个共同表达的复发性流产自噬相关基因(图1B)。30个自噬相关基因的表达水平在复发性流产患者与健康孕龄妇女之间存在明显差异,将这些差异基因绘制成热图(图1C)。为了发现这些复发性流产的自噬相关基因的潜在功能关系,利用DAVID数据库对自噬相关基因进行了GO(图1D)和KEGG(图1E)功能分析,结果显示,除了自噬相关通路外,自噬相关基因在生物过程中参与细胞分解代谢过程的调节、凋亡过程调节、病毒感染和雷帕霉素靶蛋白信号通路;在细胞组分富集分析中,自噬相关基因主要参与空泡膜的形成、线粒体或其他细胞器膜的形成等;在分子功能富集分析中,参与蛋白丝氨酸/苏氨酸激酶活性和组蛋白激酶活性等。而KEGG分析富集结果表明,自噬相关基因在人乳头瘤病毒感染、自噬调控、磷脂酰肌醇3激酶/蛋白激酶B信号通路、神经退行性变的途径等信号通路中发挥作用。 2.2 复发性流产的自噬相关基因的疾病和基因富集分析 利用Metscape对复发性流产患者的自噬相关基因进行DisGeNET富集分析。利用Metscape对复发性流产患者的自噬相关基因进行遗传功能分析,结果表明自噬相关基因与HER2基因扩增、肿瘤坏死、胰腺肿瘤、淋巴细胞和T细胞等相关(图2A)。除此之外,基因富集分析结果显示,健康对照组的相关通路主要集中于趋化因子信号通路、补体和凝血级联反应、神经活性配体受体相互作用信号(图2B),生物学功能主要集中于外部刺激的负调控过程、含有细胞外基质的胶原蛋白和刺激细胞因子的产生(图2C)。复发性流产患者的基因富集分析显示与铜离子解毒作用、金属离子的应激反应、核苷二磷酸磷酸酶活性和肌浆网等有关(图2D)。 2.3 预测风险评分模型的建立 考虑到公式中变量过多可能引发过拟合现象,并且基因间可能存在共线性问题,研究缩减了候选基因的数量,旨在降低诊断模型的偏差。图3A展示了通过LASSO回归对这些基因表达水平进行压缩的结果。通过运用LASSO回归分析与交叉验证相结合,以确定最佳的调优参数。LASSO回归不仅用于特征选择,还实现了降维,有效剔除了不重要的特征,降低了模型的复杂度。随后,对保留的特征进行了方差膨胀因子(VIF)检验,通过计算每个特征的方差膨胀因子值,量化与其他特征"
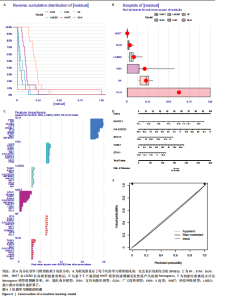
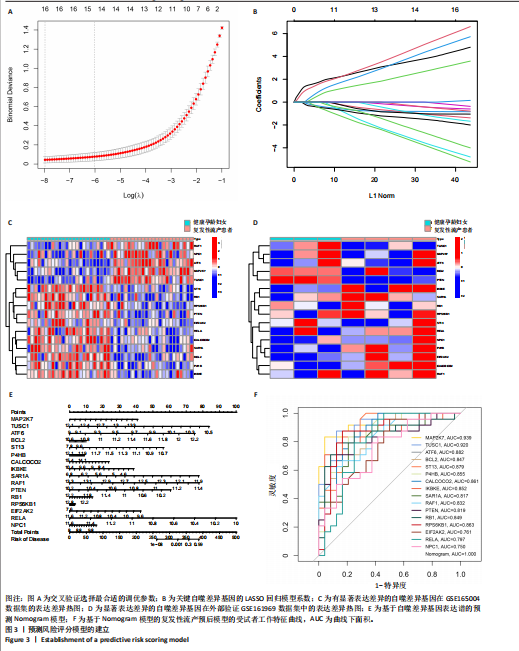
之间的共线性程度,以评估是否存在多重共线性。从图3A可以发现当log值从?8变化到?1时,偏差也随之波动。对于β系数(图3B),它们是由LASSO回归得出的,其中每条曲线代表一个基因。在剔除β系数为零的基因后,最终保留了18个基因。接下来,对这18个基因进行logistic回归分析,发现有16个关键复发性流产的自噬相关基因的P值< 0.05,并将其绘制出热图(图3C):MAP2K7、TUSC1、ATF6、BCL2、ST13、P4HB、CALCOCO2、IKBKE、SAR1A、RAF1、PTEN、RB1、RPS6KB1、EIF2AK2、RELA、NPC1。基于上述方法,利用这16个关键复发性流产的自噬相关基因构建了预测模型。为了方便诊断模型的临床使用,建立了Nomogram模型(图3D)。根据子宫内膜组织中16种复发性流产的自噬相关基因表达水平的实际测量值,可以在图中找到相应的刻度,并投影到顶部的点刻度上,读取每个变体的点,"
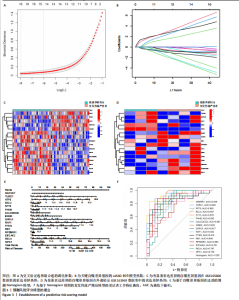
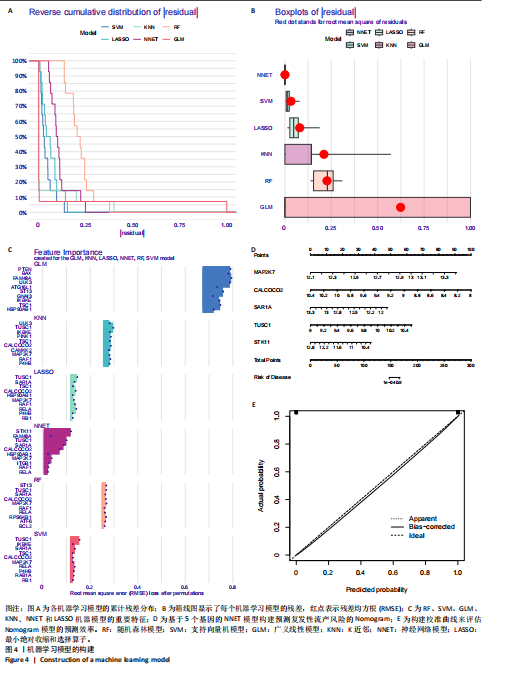
从而可以推测患复发性流产的风险概率(图3E)。通过绘制受试者工作特征曲线(图3F)来评估复发性流产诊断模型的预测准确性。 2.4 机器学习模型的构建和评估 为了进一步识别具有高诊断价值的特异性基因,基于复发性流产训练队列中特异性自噬相关基因的表达谱,建立了6种经过验证的机器学习模型(随机森林模型、支持向量机模型、广义线性模型、K近邻模型、神经网络模型和LASSO模型)。在比较以上6种不同模型时,利用贝叶斯定理来优化目标函数,通过迭代选择参数组合并更新后验分布来找到每个模型的最佳参数设置。使用“DALEX”软件包对6个模型进行解释,并绘制每个模型在测试集中的残差分布。神经网络和支持向量机模型的残差相对较低(图4A,B)。根据均方根误差(RMSE)对每个模型的前15个重要特征变量进行排序(图4C)。总的来说,结合这些结果,神经网络模型被证明可以最好地区分不同集群的患者。最后,从神经网络模型中选择前5个最重要的变量(MAP2K7、CALCOCO2、SAR1A、TUSC1和STK11)作为预测基因进行进一步分析。为了进一步评估神经网络模型的预测效率,通过构建了一个Nomogram模型来估计复发性流产患者自噬相关基因的风险(图4D)。采用校正曲线来评估Nomogram模型的预测效率。从校正曲线来看,实际的复发性流产聚类风险与预测的风险误差非常小(图4E),这些结果表明机器学习模型所构建的Nomogram模型具有较高的准确性,可以为临床决策提供依据。"
| [1] WANG J, HAN T, ZHU X. Role of maternal-fetal immune tolerance in the establishment and maintenance of pregnancy. Chin Med J (Engl). 2024;137(12):1399-1406. [2] LIN M, XU H, QIU J. Inflammation in recurrent miscarriage-a comprehensive perspective from uterine microenvironment and immune cell imbalance to therapeutic strategies. Ginekol Pol. 2023; 95(4):266-275. [3] VAN WELY M. Series of overviews on miscarriage and recurrent miscarriage. Fertil Steril. 2023;120(5):932-933. [4] BILIBIO JP, GAMA TB, NASCIMENTO ICM, et al. Causes of recurrent miscarriage after spontaneous pregnancy and after in vitro fertilization. Am J Reprod Immunol. 2020;83(5):e13226. [5] YAMAGUCHI N, TAKAKURA Y, AKIYAMA T. Autophagy and proteasomes in thymic epithelial cells: essential bulk protein degradation systems for immune homeostasis maintenance. Front Immunol. 2023;15:1488020. [6] PARZYCH KR, KLIONSKY DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Sign. 2014;20(3):460-473. [7] SHAN D, DONG R, HU Y. Current understanding of autophagy in intrahepatic cholestasis of pregnancy. Placenta. 2021;115:53-59. [8] ZHANG L, DONG Y, XU X, et al. The role of autophagy in Parkinson’s disease. Neural Regen Res. 2012;7(2):141-145. [9] QIN Q, GU Z, LI F, et al. A Diagnostic Model for Alzheimer’s Disease Based on Blood Levels of Autophagy-Related Genes. Front Aging Neurosci. 2022;14:881890. [10] GHARTEY-KWANSAH G, ADU-NTI F, ABOAGYE B, et al. Autophagy in the control and pathogenesis of parasitic infections. Cell Biosci. 2020;10:101. [11] YANG S, WANG H, LI D, et al. Role of Endometrial Autophagy in Physiological and Pathophysiological Processes. J Cancer. 2019;10(15):3459-3471. [12] ZHAO J, XU Z, XIE J, et al. The novel lnc-HZ12 suppresses autophagy degradation of BBC3 by preventing its interactions with HSPA8 to induce trophoblast cell apoptosis. Autophagy. 2024;20(10):2255-2274. [13] YE Z, MENG Q, ZHANG W, et al. Exploration of the Shared Gene and Molecular Mechanisms Between Endometriosis and Recurrent Pregnancy Loss. Front Vet Sci. 2022;9:867405. [14] XING ZY, LIU W, XING RJ, et al. Integrated analysis ceRNA network of autophagy-related gene RNF144B in steroid-induced necrosis of the femoral head. Sci Rep. 2024;14(1):28737. [15] BUSNELLI A, GAROLLA A, TERSIGNI C, et al. Sperm human papillomavirus infection and risk of idiopathic recurrent pregnancy loss: insights from a multicenter case-control study. Fertil Steril. 2023; 119(3):410-418. [16] WANG R, DAI F, DENG Z, et al. ITGA3 participates in the pathogenesis of recurrent spontaneous abortion by downregulating ULK1-mediated autophagy to inhibiting trophoblast function. Am J Physiol Cell Physiol. 2024. doi: 10.1152/ajpcell.00563.2024. [17] YANG Y, LIU B, TIAN J, et al. Vital role of autophagy flux inhibition of placental trophoblast cells in pregnancy disorders induced by HEV infection. Emerg Microbes Infect. 2023;12(2):2276336. [18] ZHAO X, JIANG Y, JIANG T, et al. Physiological and pathological regulation of autophagy in pregnancy. Arch Gynecol Obstet. 2020; 302(2):293-303. [19] SU Y, ZHANG JJ, HE JL, et al. Endometrial autophagy is essential for embryo implantation during early pregnancy. J Mol Med (Berl). 2020; 98(4):555-567. [20] DENG T, WU X, WANG Y, et al. Toe1 promotes proliferation and differentiation of neural progenitor cells. Heliyon. 2024;10(20):e39535. [21] TIAN W, LIAO H, LI N, et al. Monomethyl Phthalate Causes Early Embryo Development Delay, Apoptosis, and Energy Metabolism Disruptions Through Inducing Redox Imbalance. Reprod Sci. 2024;31(1):139-149. [22] TIAN M, ZHANG Y, LIU Z, et al. The PD-1/PD-L1 inhibitory pathway is altered in pre-eclampsia and regulates T cell responses in pre-eclamptic rats. Sci Rep. 2016;6:27683. [23] HEYDARIFARD Z, ZADHEIDAR S, YAVARIAN J, et al. Potential role of viral infections in miscarriage and insights into the underlying molecular mechanisms. Congenit Anom (Kyoto). 2022;62(2):54-67. [24] ALNAES-KATJAVIVI P, ROALD B, STAFF AC. Uteroplacental acute atherosis in preeclamptic pregnancies: Rates and clinical outcomes differ by tissue collection methods. Pregnancy Hypertens. 2020;19:11-17. [25] MATSUZAKI S, GREMEAU AS, POULY JL. Impaired pathogen-induced autophagy and increased IL-1β and TNFα release in response to pathogenic triggers in secretory phase endometrial stromal cells of endometriosis patients. Reprod Biomed Online. 2020;41(5):767-781. [26] LIU J, CHAKRABORTY C, GRAHAM CH, et al. Noncatalytic domain of uPA stimulates human extravillous trophoblast migration by using phospholipase C, phosphatidylinositol 3-kinase and mitogen-activated protein kinase. Exp Cell Res. 2003;286(1):138-151. [27] LIU J, PUSCHECK EE, WANG F, et al. Serine-threonine kinases and transcription factors active in signal transduction are detected at high levels of phosphorylation during mitosis in preimplantation embryos and trophoblast stem cells. Reproduction. 2004;128(5):643-654. [28] BRUNO MT, CARUSO S, SCALIA G, et al. Papillomavirus Infection as Potential Cause of Miscarriage in the Early Gestational Age: A Prospective Study. Diagnostics (Basel). 2023;13(9):1659. [29] MA N, LIU B, JIN Y, et al. Aquaporin 9 causes recurrent spontaneous abortion by inhibiting trophoblast cell epithelial-mesenchymal transformation and invasion through the PI3K/AKT pathway. Biol Reprod. 2023;109(5):736-748. [30] SUN C, ROSENSTOCK TR, COHEN MA, et al. Autophagy Dysfunction as a Phenotypic Readout in hiPSC-Derived Neuronal Cell Models of Neurodegenerative Diseases. Methods Mol Biol. 2022;2549:103-136. [31] RANA T, BEHL T, SEHGAL A, et al. Exploring the Role of Autophagy Dysfunction in Neurodegenerative Disorders. Mol Neurobiol. 2021; 58(10):4886-4905. [32] BOUDY AS, FERRIER C, SELLERET L, et al. Prognosis of HER2-positive pregnancy-associated breast cancer: Analysis from the French CALG (Cancer Associé à La Grossesse) network. Breast. 2020;54:311-318. [33] RAY D, YUN YC, IDRIS M, et al. A tumor-associated splice-isoform of MAP2K7 drives dedifferentiation in MBNL1-low cancers via JNK activation. Proc Natl Acad Sci U S A. 2020;117(28):16391-16400. [34] HE S, NING Y, MA F, et al. IL-23 Inhibits Trophoblast Proliferation, Migration, and EMT via Activating p38 MAPK Signaling Pathway to Promote Recurrent Spontaneous Abortion. J Microbiol Biotechnol. 2022;32(6):792-799. [35] HAM J, SONG J, SONG G, et al. Oryzalin impairs maternal-fetal interaction during early pregnancy via ROS-mediated P38 MAPK/AKT and OXPHOS downregulation. Food Chem Toxicol. 2023;174:113665. [36] SANG W, YAN X, WANG L, et al. CALCOCO2 prevents AngII-induced atrial remodeling by regulating the interaction between mitophagy and mitochondrial stress. Int Immunopharmacol. 2024;140:112841. [37] YAMANO K, YOULE RJ. Two different axes CALCOCO2-RB1CC1 and OPTN-ATG9A initiate PRKN-mediated mitophagy. Autophagy. 2020; 16(11):2105-2107. [38] YAN C, GONG L, CHEN L, et al. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy. 2020;16(3):419-434. [39] ZHAO Y, LIANG Y, CAI L, et al. Comprehensive Proteomic Analysis Reveals Distinct Features and a Diagnostic Biomarker Panel for Early Pregnancy Loss in Histological Subtypes. Mol Cell Proteomics. 2024;23(11):100848. [40] XIAO J, LI W, LI G, et al. STK11 overexpression prevents glucocorticoid-induced osteoporosis via activating the AMPK/SIRT1/PGC1α axis. Hum Cell. 2022;35(4):1045-1059. [41] HUANG Y, ZHANG H, FENG J, et al. STK11 mutation affects the killing effect of NK cells to promote the progression of lung adenocarcinoma. APMIS. 2022;130(11):647-656. [42] ALHARBI KK, KHAN IA, ELDESOUKY MH, et al. The genetic polymorphism in the STK11 does not affect gestational diabetes. Acta Biochim Pol. 2015;62(3):569-572. |
| [1] | Jia Jinwen, Airefate·Ainiwaer, Zhang Juan. Effects of EP300 on autophagy and apoptosis related to allergic rhinitis in rats [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1439-1449. |
| [2] | Lai Jiaming, , Song Yuling, Chen Zixi, Wei Jinghuan, Cai Hao, , Li Guoquan, . Screening of diagnostic markers for endothelial cell Senescence in mice with radiation-induced heart disease and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1450-1463. |
| [3] | You Huijuan, Wu Shuzhen, Rong Rong, Chen Liyuan, Zhao Yuqing, Wang Qinglu, Ou Xiaowei, Yang Fengying. Macrophage autophagy in lung diseases: two-sided effects [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1516-1526. |
| [4] | Zhang Qian, Huang Dongfeng. Weighted gene co-expression network analysis combined with machine learning to screen and validate biomarkers for osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1096-1105. |
| [5] | Liu Kexin, , Hao Kaimin, Zhuang Wenyue, , Li Zhengyi. Autophagy-related gene expression in pulmonary fibrosis models: bioinformatic analysis and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1129-1138. |
| [6] | Hu Jing, Zhu Ling, Xie Juan, Kong Deying, Liu Doudou. Autophagy regulates early embryonic development in mice via affecting H3K4me3 modification [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1147-1155. |
| [7] | Gu Fucheng, Yang Meixin, Wu Weixin, Cai Weijun, Qin Yangyi, Sun Mingyi, Sun Jian, Geng Qiudong, Li Nan. Effects of Guilu Erxian Glue on gut microbiota in rats with knee osteoarthritis: machine learning and 16S rDNA analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1058-1072. |
| [8] | Guan Yujie, Zhao Bin. Application and prospect of artificial intelligence in screening and diagnosis of scoliosis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 721-730. |
| [9] | Wang Zhipeng, Zhang Xiaogang, Zhang Hongwei, Zhao Xiyun, Li Yuanzhen, Guo Chenglong, Qin Daping, Ren Zhen. A systematic review of application value of machine learning to prognostic prediction models for patients with lumbar disc herniation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 740-748. |
| [10] | Zhao Feifan, Cao Yujing. An artificial neural network model of ankylosing spondylitis and psoriasis shared genes and machine learning-based mining and validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 770-784. |
| [11] | Liu Chu, Qiu Boyuan, Tong Siwen, He Linyuwei, Chen Haobo, Ou Zhixue. A genetic perspective reveals the relationship between blood metabolites and osteonecrosis: an analysis of information from the FinnGen database in Finland [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 785-794. |
| [12] | Hu Yalin, Huang Fengqin, Yang Boyin, Luo Xingmei. Transcription factor EB improves Alzheimer’s disease via the autophagy-lysosome pathway [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(22): 5844-5858. |
| [13] | Zheng Wen, Zhu Dongsheng, Wang Xiaodong. Secreted modular calcium binding protein regulates autophagy in the acetabular cartilage of rats with developmental dysplasia of the hip [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(18): 4618-4626. |
| [14] | Zhang Shuli, Hou Chaowen, Yuan Shanshan, Ma Yuhua . Mechanism by which exercise regulates autophagy in different physiological systems [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(18): 4737-4748. |
| [15] | Cheng Yuebin, Wang Baojian, Dai Wenkang, Yin Yueshan, Sun Zhiqiang, Peng Zhiyun, Shang Yuhang, Ma Yufeng. Revealing the regulatory targets of plasma proteins in rheumatoid arthritis from the perspective of genomics [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(17): 4498-4507. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||