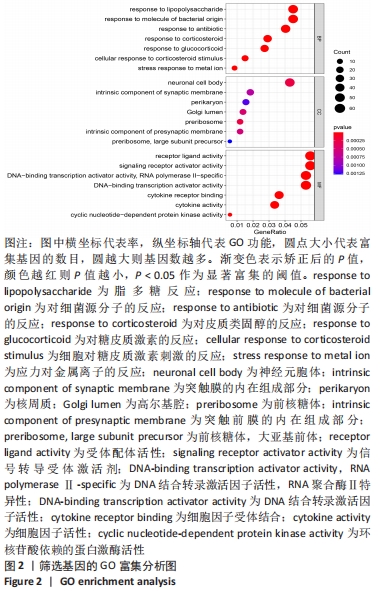
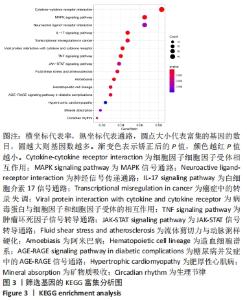
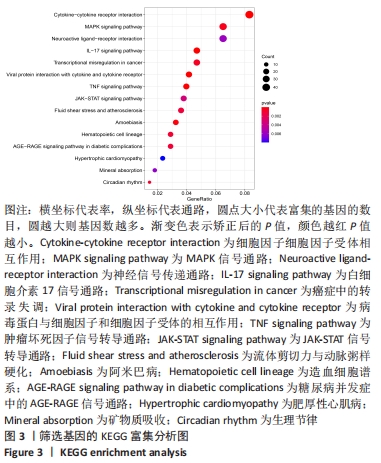
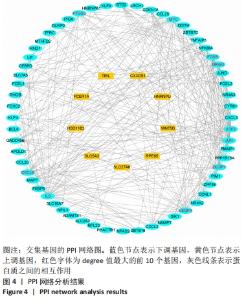
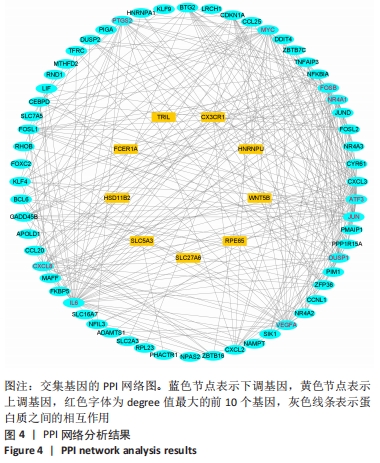
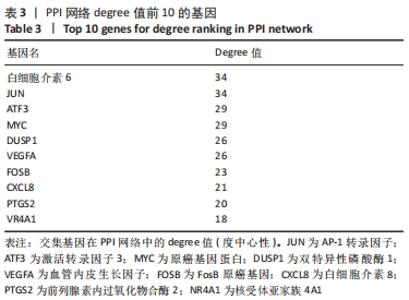
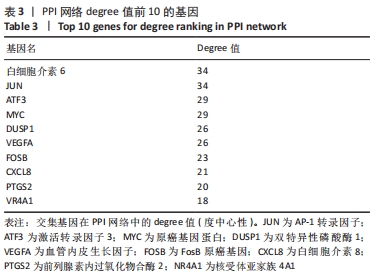
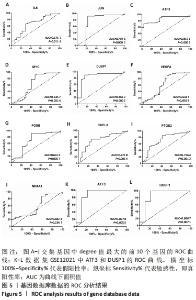
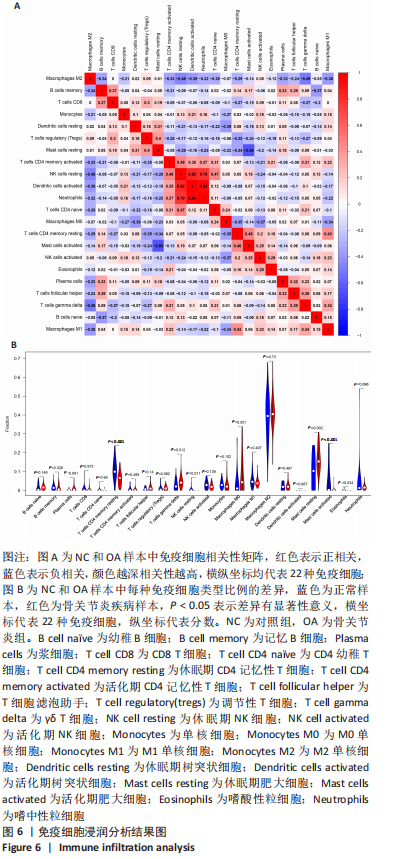
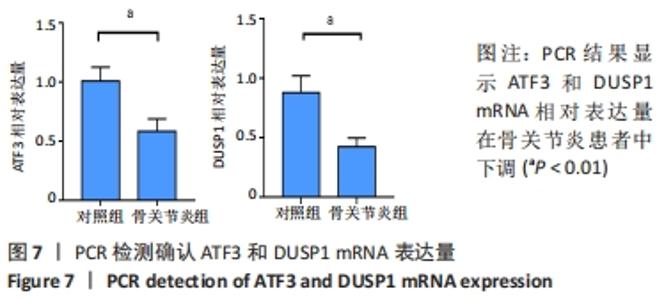
Chinese Journal of Tissue Engineering Research ›› 2022, Vol. 26 ›› Issue (33): 5342-5349.doi: 10.12307/2022.952
Previous Articles Next Articles
Screening key genes in synovium of osteoarthritis by a combination of differentially expressed genes and weighted co-expression network analysis
Qian Xiaofen1, Zeng Ping2, Liu Jinfu1, Wang Hao1, Zhou Shulong1, Pan Haida1
- 1School of Graduate, Guangxi University of Chinese Medicine, Nanning 530299, Guangxi Zhuang Autonomous Region, China; 2Second Department of Orthopedics, Xianhu Branch, First Affiliated Hospital, Guangxi University of Chinese Medicine, Nanning 530023, Guangxi Zhuang Autonomous Region, China
-
Received:2021-03-23Accepted:2021-04-28Online:2022-11-28Published:2022-03-31 -
Contact:Zeng Ping, MD, Chief physician, Second Department of Orthopedics, Xianhu Branch, First Affiliated Hospital, Guangxi University of Chinese Medicine, Nanning 530023, Guangxi Zhuang Autonomous Region, China -
About author:Qian Xiaofen, Master candidate, School of Graduate, Guangxi University of Chinese Medicine, Nanning 530299, Guangxi Zhuang Autonomous Region, China -
Supported by:Chinese Medicine Appropriate Technology Development and Promotion Project of Guangxi Zhuang Autonomous Region Traditional Chinese Medicine Bureau, No. GZSY21-14 (to ZP)
CLC Number:
Cite this article
Qian Xiaofen, Zeng Ping, Liu Jinfu, Wang Hao, Zhou Shulong, Pan Haida. Screening key genes in synovium of osteoarthritis by a combination of differentially expressed genes and weighted co-expression network analysis[J]. Chinese Journal of Tissue Engineering Research, 2022, 26(33): 5342-5349.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
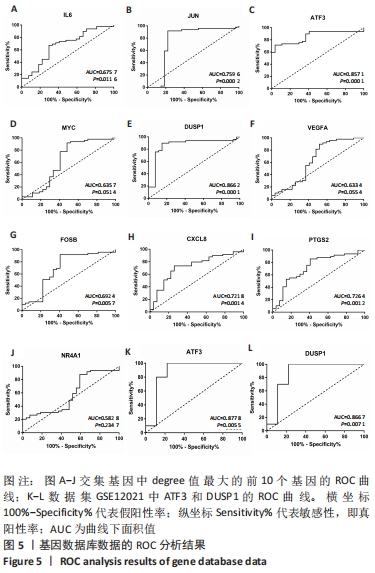
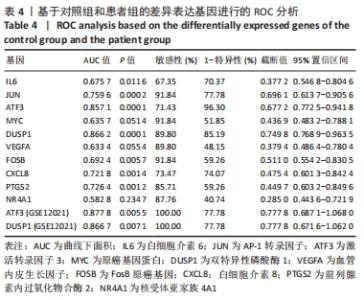
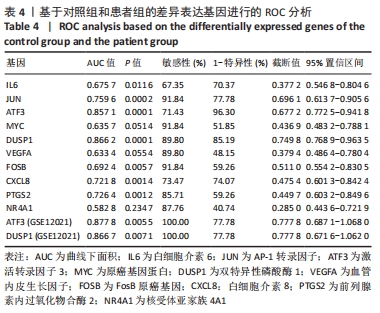
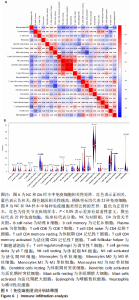
| [1] CHEN H, CHEN L. An integrated analysis of the competing endogenous RNA network and co-expression network revealed seven hub long non-coding RNAs in osteoarthritis. Bone Joint Res. 2020;9(3):90-98. [2] HE Y, LI Z, ALEXANDER PG, et al. Pathogenesis of osteoarthritis: risk factors, regulatory pathways in chondrocytes, and experimental models. Biology (Basel). 2020;9(8):194. [3] HAN D, FANG Y, TAN X, et al. The emerging role of fibroblast-like synoviocytes-mediated synovitis in osteoarthritis: an update. J Cell Mol Med. 2020;24(17):9518-9532. [4] MATHIESSEN A, CONAGHAN PG. Synovitis in osteoarthritis: current understanding with therapeutic implications. Arthritis Res Ther. 2017; 19(1):18. [5] CHEN H, WANG X, JIA H, et al. Bioinformatics analysis of key genes and pathways of cervical cancer. Onco Targets Ther. 2020;13:13275-13283. [6] XIU MX, LIU YM, CHEN GY, et al. Identifying Hub Genes, Key Pathways and Immune Cell Infiltration Characteristics in Pediatric and Adult Ulcerative Colitis by Integrated Bioinformatic Analysis. Dig Dis Sci. 2020. doi: 10.1007/s10620-020-06611-w. [7] ZHU Z, ZHONG L, LI R, et al. Study of osteoarthritis-related hub genes based on bioinformatics analysis. Biomed Res Int. 2020;2020:2379280. [8] LI Z, ZHONG L, DU Z, et al. Network analyses of differentially expressed genes in osteoarthritis to identify hub genes. Biomed Res Int. 2019; 2019:8340573. [9] WANG Q, ROZELLE AL, LEPUS CM, et al. Identification of a central role for complement in osteoarthritis. Nat Med. 2011;17(12):1674-1679. [10] WOETZEL D, HUBER R, KUPFER P, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther. 2014;16(2):R84. [11] BROEREN MG, DE VRIES M, BENNINK MB, et al. Functional tissue analysis reveals successful cryopreservation of human osteoarthritic synovium. PLoS One. 2016;11(11):e0167076. [12] QIAN S, SUN S, ZHANG L, et al. Integrative analysis of dna methylation identified 12 signature genes specific to metastatic ccRCC. Front Oncol. 2020;10:556018. [13] LI G, LI X, YANG M, et al. Prediction of biomarkers of oral squamous cell carcinoma using microarray technology. Sci Rep. 2017;7:42105. [14] AKI T, HASHIMOTO K, OGASAWARA M, et al. A whole-genome transcriptome analysis of articular chondrocytes in secondary osteoarthritis of the hip. PLoS One. 2018;13(6):e0199734. [15] SZKLARCZYK D, FRANCESCHINI A, WYDER S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447-D452. [16] JAYADEV C, HULLEY P, SWALES C, et al. Synovial fluid fingerprinting in end-stage knee osteoarthritis: a novel biomarker concept. Bone Joint Res. 2020;9(9):623-632. [17] HUBER R, HUMMERT C, GAUSMANN U, et al. Identification of intra-group, inter-individual, and gene-specific variances in mRNA expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther. 2008;10(4):R98. [18] KANG K, XIE F, MAO J, et al. Significance of tumor mutation burden in immune infiltration and prognosis in cutaneous melanoma. Front Oncol. 2020;10:573141. [19] 骨关节炎诊疗指南(2018年版)[J].中华骨科杂志,2018,38(12):705-715. [20] CALABRESE G, ARDIZZONE A, CAMPOLO M, et al. Beneficial effect of tempol, a membrane-permeable radical scavenger, on inflammation and osteoarthritis in in vitro models. Biomolecules. 2021;11(3):352. [21] CAI W, LI H, ZHANG Y, et al. Identification of key biomarkers and immune infiltration in the synovial tissue of osteoarthritis by bioinformatics analysis. PeerJ. 2020;8:e8390. [22] ZHAO Q, WANG Q, XU J, et al. Expression of KCNA2 in the dorsal root ganglia of rats with osteoarthritis pain induced by monoiodoacetate. Nan Fang Yi Ke Da Xue Xue Bao. 2019;39(5):579-585. [23] IEZAKI T, OZAKI K, FUKASAWA K, et al. ATF3 deficiency in chondrocytes alleviates osteoarthritis development. J Pathol. 2016;239(4):426-437. [24] SHIN SM, CAI Y, ITSON-ZOSKE B, et al. Enhanced T-type calcium channel 3.2 activity in sensory neurons contributes to neuropathic-like pain of monosodium iodoacetate-induced knee osteoarthritis. Mol Pain. 2020;16:1744806920963807. [25] GONÇALVES DOS SANTOS G, JIMENÉZ-ANDRADE JM, WOLLER SA, et al. The neuropathic phenotype of the K/BxN transgenic mouse with spontaneous arthritis: pain, nerve sprouting and joint remodeling. Sci Rep. 2020;10(1):15596. [26] BLOM AB, VAN DEN BOSCH MH, BLANEY DAVIDSON EN, et al. The alarmins S100A8 and S100A9 mediate acute pain in experimental synovitis. Arthritis Res Ther. 2020;22(1):199. [27] NUNES-XAVIER C, ROMÁ-MATEO C, RÍOS P, et al. Dual-specificity MAP kinase phosphatases as targets of cancer treatment. Anticancer Agents Med Chem. 2011;11(1):109-132. [28] HOPPSTÄDTER J, AMMIT AJ. Role of Dual-specificity phosphatase 1 in glucocorticoid-driven anti-inflammatory responses. Front Immunol. 2019;10:1446. [29] CHOI Y, YOO JH, LEE Y, et al. Calcium-phosphate crystals promote RANKL expression via the downregulation of DUSP1. Mol Cells. 2019; 42(2):183-188. [30] PENG HZ, YUN Z, WANG W, et al. Dual specificity phosphatase 1 has a protective role in osteoarthritis fibroblast‑like synoviocytes via inhibition of the MAPK signaling pathway. Mol Med Rep. 2017;16(6):8441-8447. [31] 彭华志.DUSP1在人正常及骨关节炎滑膜细胞中的表达及对骨关节炎的保护作用[D].西安:第四军医大学,2017. [32] LI W, ZHANG Z, WANG ZM. Differential immune cell infiltrations between healthy periodontal and chronic periodontitis tissues. BMC Oral Health. 2020;20(1):293. [33] ROHR-UDILOVA N, KLINGLMÜLLER F, SCHULTE-HERMANN R, et al. Deviations of the immune cell landscape between healthy liver and hepatocellular carcinoma. Sci Rep. 2018;8(1):6220. [34] CHEN B, KHODADOUST MS, LIU CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243-259. [35] CHEN Z, MA Y, LI X, et al. The immune cell landscape in different anatomical structures of knee in osteoarthritis: a gene expression-based study. Biomed Res Int. 2020;2020:9647072. |
| [1] | Zhang Jichao, Dong Yuefu, Mou Zhifang, Zhang Zhen, Li Bingyan, Xu Xiangjun, Li Jiayi, Ren Meng, Dong Wanpeng. Finite element analysis of biomechanical changes in the osteoarthritis knee joint in different gait flexion angles [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1357-1361. |
| [2] | Jin Tao, Liu Lin, Zhu Xiaoyan, Shi Yucong, Niu Jianxiong, Zhang Tongtong, Wu Shujin, Yang Qingshan. Osteoarthritis and mitochondrial abnormalities [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(9): 1452-1458. |
| [3] | Wang Baojuan, Zheng Shuguang, Zhang Qi, Li Tianyang. Miao medicine fumigation can delay extracellular matrix destruction in a rabbit model of knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(8): 1180-1186. |
| [4] | Gao Yujin, Peng Shuanglin, Ma Zhichao, Lu Shi, Cao Huayue, Wang Lang, Xiao Jingang. Osteogenic ability of adipose stem cells in diabetic osteoporosis mice [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(7): 999-1004. |
| [5] | Liu Dongcheng, Zhao Jijun, Zhou Zihong, Wu Zhaofeng, Yu Yinghao, Chen Yuhao, Feng Dehong. Comparison of different reference methods for force line correction in open wedge high tibial osteotomy [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(6): 827-831. |
| [6] | Zhou Jianguo, Liu Shiwei, Yuan Changhong, Bi Shengrong, Yang Guoping, Hu Weiquan, Liu Hui, Qian Rui. Total knee arthroplasty with posterior cruciate ligament retaining prosthesis in the treatment of knee osteoarthritis with knee valgus deformity [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(6): 892-897. |
| [7] | He Junjun, Huang Zeling, Hong Zhenqiang. Interventional effect of Yanghe Decoction on synovial inflammation in a rabbit model of early knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 694-699. |
| [8] | Lin Xuchen, Zhu Hainian, Wang Zengshun, Qi Tengmin, Liu Limin, Suonan Angxiu. Effect of xanthohumol on inflammatory factors and articular cartilage in a mouse mode of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 676-681. |
| [9] | Liu Jin, Li Zhen, Hao Huiqin, Wang Ze, Zhao Caihong, Lu Wenjing. Ermiao san aqueous extract regulates proliferation, migration, and inflammatory factor expression of fibroblast-like synovial cells in collagen-induced arthritis rats [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 688-693. |
| [10] | Xu Lei, Han Xiaoqiang, Zhang Jintao, Sun Haibiao. Hyaluronic acid around articular chondrocytes: production, transformation and function characteristics [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 768-773. |
| [11] | Li Jiajun, Xia Tian, Liu Jiamin, Chen Feng, Chen Haote, Zhuo Yinghong, Wu Weifeng. Molecular mechanism by which icariin regulates osteogenic signaling pathways in the treatment of steroid-induced avascular necrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(5): 780-785. |
| [12] | Zhang Tong, Cai Jinchi, Yuan Zhifa, Zhao Haiyan, Han Xingwen, Wang Wenji. Hyaluronic acid-based composite hydrogel in cartilage injury caused by osteoarthritis: application and mechanism [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(4): 617-625. |
| [13] | Song Wei, Zhang Yaxin, Jia Dazhou, Sun Yu. Adjective application of dexamethasone combined with furosemide for early pain and swelling after total knee arthroplasty [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(33): 5317-5322. |
| [14] | Li Yi, Yang Yanjun, Peng Songyun, Cheng Zhigang, Zhong Kai, Yin Tianping, Tang Lianghua. Effect of Miao medicine Jiuxian Luohan Jiegu Decoction on osteogenic differentiation and fracture healing in tibial fracture rats [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(33): 5350-5356. |
| [15] | Huang Wei, Dong Panfeng, Huang Yourong, Xia Tian. Epimedium in regulating bone marrow mesenchymal stem cell differentiation and preventing osteoporosis related signaling pathways [J]. Chinese Journal of Tissue Engineering Research, 2022, 26(30): 4889-4895. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||