Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (13): 3446-3457.doi: 10.12307/2026.204
Previous Articles Next Articles
Hot spot and current status of single-cell RNA sequencing in stroke field
Cao Lifang1, Chen Tao1, Shou Jiayin1, Fan Fangfang1, 2
- 1Guizhou Medical University, Gui'an New Area 561113, Guizhou Province, China;2Affiliated Hospital of Guizhou Medical University, Guiyang 550025, Guizhou Province, China
-
Accepted:2025-09-11Online:2026-05-08Published:2025-12-26 -
Contact:Fan Fangfang, PhD, Associate professor, Master's supervisor, Guizhou Medical University, Gui'an New Area 561113, Guizhou Province, China; Affiliated Hospital of Guizhou Medical University, Guiyang 550025, Guizhou Province, China -
About author:Cao Lifang, Guizhou Medical University. Gui'an New Area 561113, Guizhou Province, China -
Supported by:National Natural Science Foundation of China, No. 82460781 (to FFF); China Postdoctoral Science Foundation, No. 2022MD723771 (to FFF)
CLC Number:
Cite this article
Cao Lifang, Chen Tao, Shou Jiayin, Fan Fangfang. Hot spot and current status of single-cell RNA sequencing in stroke field[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(13): 3446-3457.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
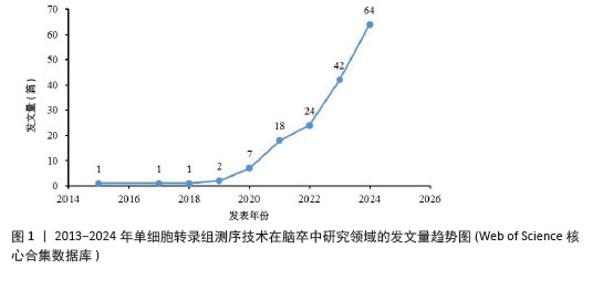
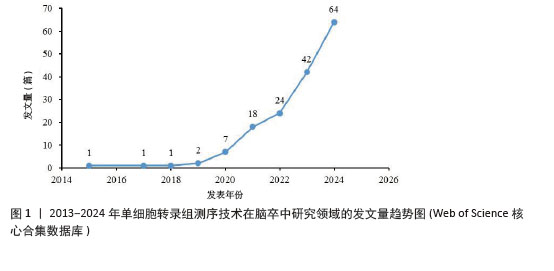
2.1 2013-2024年度发文量及时间分布分析 从近10年的发文量数据来看(图1),单细胞转录组测序技术在脑卒中研究领域的发文量逐年上升,2024年高达64篇,提示单细胞转录组测序技术越来越受到该领域科研人员的关注,这可能得益于单细胞转录组测序技术本身的不断发展和完善,使它在研究脑卒中这一复杂疾病时更具可行性和有效性。同时,这也预示着未来该领域可能会有更多的研究成果涌现,进而促进对脑卒中发病机制的认识和治疗方法的探索。其中,2019年后发文量上升速度加快,可能与全球范围内对神经系统疾病研究投入的增加以及单细胞转录组测序技术在这一时期取得的关键进展有关,使得更多研究团队能够开展相关研究。 "
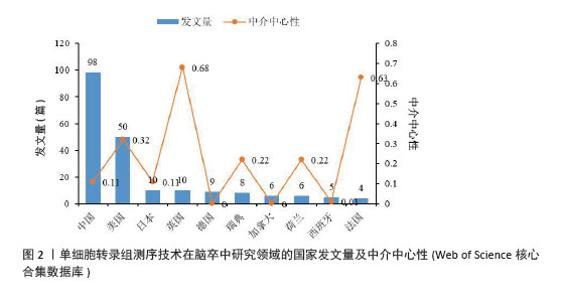
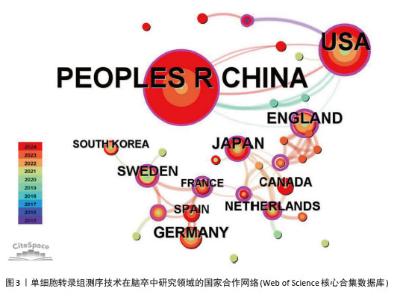
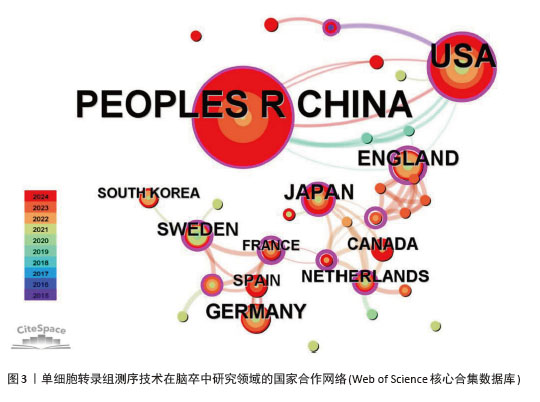
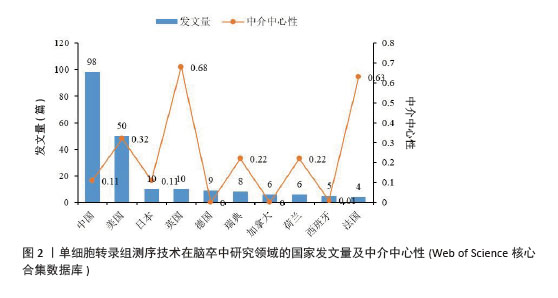
2.2 国家和机构分析 2.2.1 国家(地区)合作网络 文献分析结果显示,共有33个国家在脑卒中领域进行单细胞转录组测序的研究,其中中国(98篇)和美国(50篇)发文量较多,紧随其后的是英国(10篇)、日本(10篇)和德国(9篇),见图2。国家合作网络中有节点33个,连线62条,密度为0.117 4(连线颜色代表两者第一次合作年份,粗细则表示两者的合作频次),见图3。中国和美国成为发文量较多的2个国家,反映出两国在生命科学研究领域的强大实力和对脑卒中研究领域的重视。中国虽然发文量领先,但中心中介性相对较低,说明在国际合作方面还有提升空间。中国研究团队应积极寻求与其他国家的合作,融入国际科研网络,共享研究成果和经验,从而提升中国在该领域研究的影响力。英国、法国等国虽然发文量相对较少,但中介中心性较高,表明他们在国际合作中扮演着重要的角色。中国研究机构或可以借鉴这些国家的合作模式,加强与国际同行的交流与合作,拓展研究视野,深入开展脑卒中领域的相关研究。"
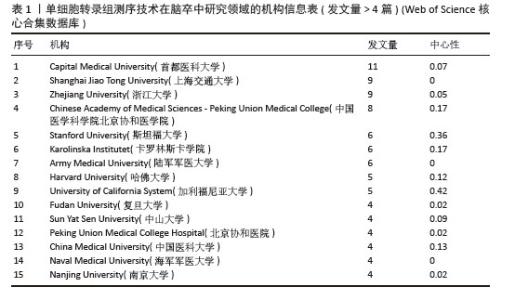
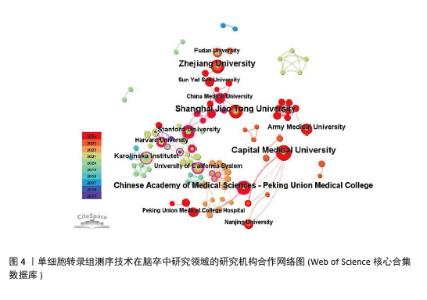
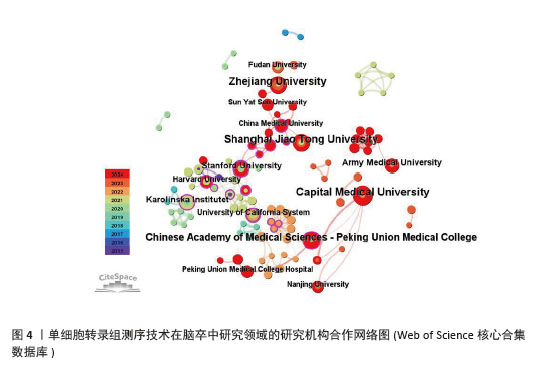
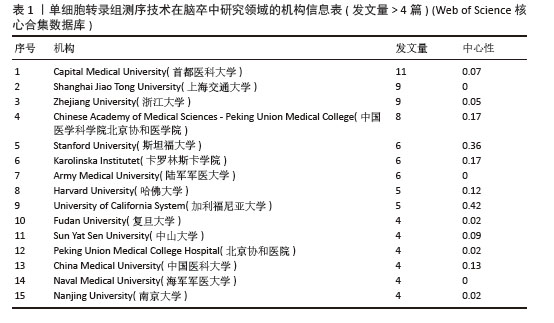
2.2.2 机构合作网络 Web of Science核心合集数据库中发文量4篇以上的研究机构见表1。排名前3的分别是首都医科大学、上海交通大学、浙江大学。其中首都医科大学的中介中心性最高,表明与其他单位(布里格姆&欧姆恩施医院、中国科学院)合作密切。机构共现关系图谱中节点有108个,连线数186条,密度为0.031 6,证明共有108家机构被纳入图谱且各个机构之间合作紧密,见图4。近年发文量较多的机构主要来自于中国(首都医科大学、上海交通大学、浙江大学)和美国(斯坦福大学)。首都医科大学、哈佛大学等机构的高发文量和积极的合作态势表明,这些机构具备先进的研究设施和优秀的科研团队,能够在该领域持续开展深入研究。其他机构可以学习他们的科研管理经验,加强学科建设和人才培养,提升自身在该领域的研究能力。"
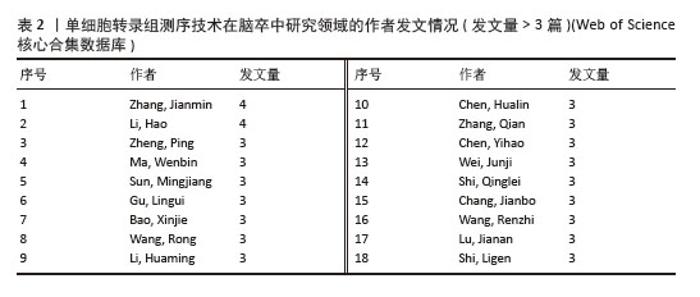
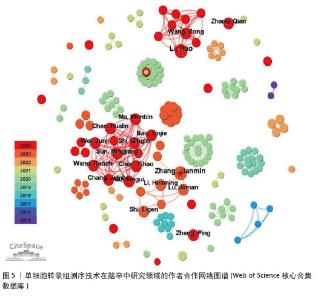
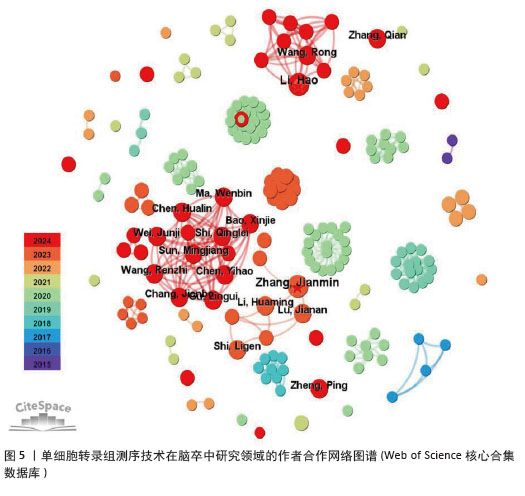
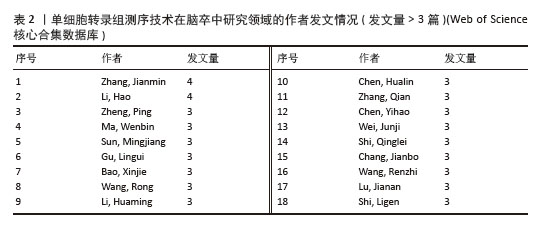
2.3 作者合作网络分析 根据普赖斯定律[26],计算得出发文量大于2(1.498)篇的核心作者共计60位,发文量3篇以上的作者见表2,排在首位的作者是Zhang,Jianmin(4篇)(兰州大学)。在作者合作网络图谱中(图5),节点数为194个,连线数为503条,密度为0.026 9,表示共有194位作者被纳入图谱中。核心作者之间的合作网络可以促进知识共享和技术交流,提高研究效率。以张建民团队为例,其在该领域的研究方向聚焦于小胶质细胞与巨噬细胞在神经炎症中的作用,应用单细胞转录组测序技术有助于深入挖掘细胞层面的疾病机制,为后续研究提供了更明确的方向。未来,随着应用研究的深入,可能会出现更多跨机构、跨学科的合作团队,整合不同领域的专业知识,为解决脑卒中这一复杂疾病带来新的突破。"
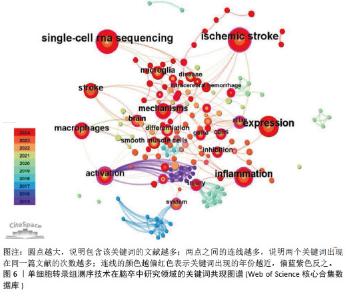
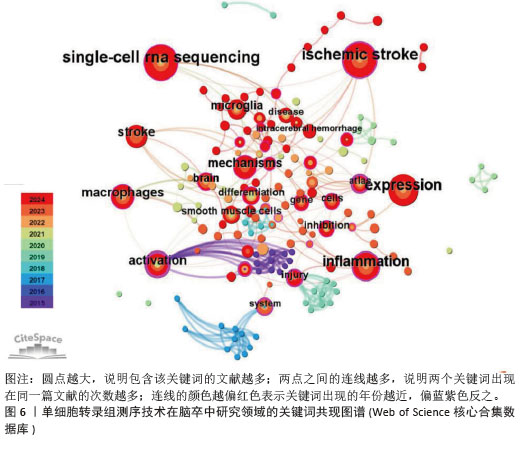
2.4 关键词共现分析 关键词共现图谱可直观地体现该领域的研究主题,使读者快速了解文献内容。在图6中,节点数为157个,连线数为310条,表示共有个157关键词纳入图谱中(圆点越大,说明包含该关键词的文献越多;两点之间的连线越多,说明两个关键词出现在同一篇文献的次数越多;连线的颜色越偏红色表示关键词出现的年份越近,偏蓝紫色反之)。在关键词共现图谱中,中介中心性≥0.1说明其中介中心性较高,而中心中介性较高的节点通常为连接两个领域的关键枢纽,经常被用作衡量文献的重要性指标。在关键词共现分析中,中心中介性≥0.1的节点有28个,≥0.3的节点有3个,分别是activation(0.63)、atlas(0.52)、dynamics(0.4)。"
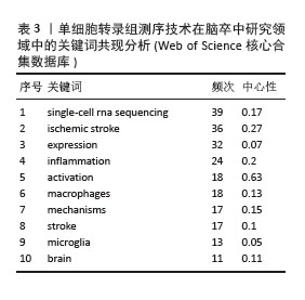
按关键词出现频数由高到低排列(表3),排名前10的分别是:single-cell rna sequencing(单细胞RNA测序),ischemic stroke(缺血性脑卒中),expression(表达),inflammation (炎症),activation(激活),macrophages (巨噬细胞),mechanisms(机制)、stroke(脑卒中),microglia(小胶质细胞),brain(大脑)。此数据提示单细胞转录组测序技术在脑卒中领域研究以缺血性脑卒中为主,侧重从炎症的角度揭示巨噬细胞、小胶质细胞等免疫细胞激活或表达对脑卒中病程的影响突出了当前研究热点可能主要集中在监测小胶质细胞或巨噬细胞,以探索炎症反应在该疾病进程中的发病机制。"
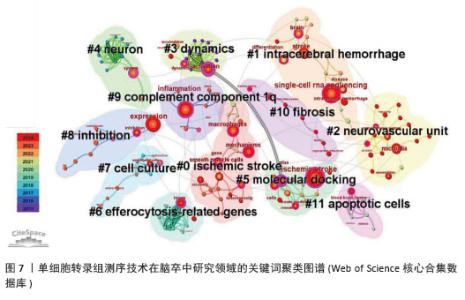
2.5 关键词聚类及时间线分析 如图7所示聚类图谱,能够直观地反映出单细胞转录组测序技术在脑卒中研究领域的多个主题。图中共得到12个聚类,聚类后得出的视图中显示模块值Q=0.800 6 > 0.3,平均轮廓值S=0.909 7 > 0.7,证明其得出的结构显著,结果高效。图中的每个色块代表一个聚类,编号越小代表所包含的成员数量越多。通过LLR算法得出关键词聚类标签列表,主要包括#0缺血性脑卒中、#1 intracerebral hemorrhage(脑出血)、#2 neurovascular unit(神经血管单元)、#3 dynamics(动力学)、#4 neuron(神经元)、#5 molecular docking(分子对接)、#6 efferocytosis-related genes(胞葬相关基因)、#7 cell culture(细胞培养)、#8 inhibition(抑制)、#9 complement component1q(补体成分1q,C1q)、#10 fibrosis(纤维化)和#11 apoptotic cells(凋亡细胞)。由此可见,单细胞转录组测序技术研究涉及缺血性和出血性脑卒中,包含神经元、神经血管单元等相关细胞,结合细胞培养和分子对接技术,研究脑卒中后抑制细胞相关功能基因(如胞葬相关基因),从细胞动力学、纤维化、补体成分1q和细胞凋亡等多方面揭示脑卒中病理变化的影响因素。"
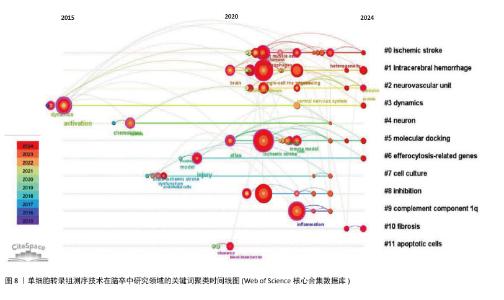
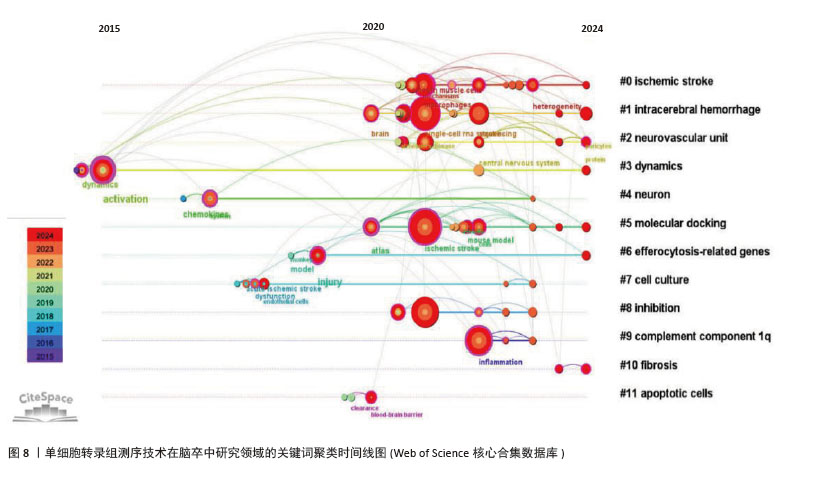
关键词聚类时间线图谱中(图8),相同聚类的关键词放置在同一水平线上,越靠右时间越近,图中的圆形节点代表研究关键词,节点越大,表明该关键词出现的频次越高、越受关注。节点之间的连线表示关键词之间的关联,连线越粗,关联越紧密。从图中节点分布情况可知,各大节点主要聚集在2020年以后。在聚类#1脑出血中,利用单细胞RNA测序技术阐明了炎症与细胞异质性的交互关系:炎症微环境通过影响细胞基因表达塑造异质性(如促炎因子诱导星形胶质细胞向A1型转化),而细胞异质性又通过分泌特定因子反作用于炎症进程(如修复型小胶质细胞分泌白细胞介素10抑制过度炎症),明确了二者交互的分子路径,为开发兼顾抗炎与调节细胞异质性的治疗药物提供了科学依据[10]。借助单细胞转录组测序技术,脑出血研究实现了细胞层面的精细解析,如在脑出血微环境中,神经元、星形胶质细胞、少突胶质细胞、小胶质细胞及浸润的免疫细胞等均表现出高度异质性[27]。分子对接作为计算生物学技术,通过模拟药物分子与靶点蛋白的结合模式,可以预测治疗神经疾病药物的作用机制。聚类#5“molecular docking”(分子对接)与 “ischemic stroke”“intracerebral hemorrhage” 等疾病聚类关联,表明分子对接技术在出血性或缺血性脑卒中的单细胞测序技术研究文献中有应用,追溯文献发现,SUN等[28]以苓桂术甘汤(LZD)为研究对象,基于网络药理学筛选的苓桂术甘汤活性成分(如槲皮素、山柰酚),通过分子对接(AutoDock Vina)验证其与炎症核心靶点(肿瘤坏死因子、白细胞介素6)的高亲和力结合,从分子层面揭示其通过抑制肿瘤坏死因子/核因子κB通路及氧化应激的分子互作基础;进一步结合单细胞测序技术分析,发现苓桂术甘汤靶基因在促修复细胞亚群(如M2巨噬细胞、内皮细胞)中特异性富集,并通过基因集变异分析(GSVA)阐明其调控M1/M2巨噬细胞极化、促进内皮修复的细胞动态,抑制神经炎症并改善脑缺血损伤。分子对接锁定靶点互作,单细胞测序定位细胞功能网络,不仅验证了中药复方多靶点协同作用的科学性,还为脑卒中治疗提供了从分子机制到细胞异质性的精准干预策略,凸显多组学整合在解析复杂疾病机制中的独特优势。研究发现,在脑出血、缺血性脑卒中等疾病中,异常表达的补体成分1q可激活补体系统,加剧神经炎症,促进神经元凋亡[29]。聚类#9“complement component1q”(补体成分1q,C1q)与“inflammation”(炎症)、“neuronal apoptosis”(神经元凋亡)等节点关联,提示补体成分1q可能通过调控炎症和细胞凋亡影响神经疾病进展。2024年 ,一项研究通过野生型(WT)与 C1qa 基因敲除(KO)小鼠构建脑出血模型,利用MRI、免疫组化分析脑损伤机制,并注射凝血酶(脑缺血损伤关键因子)进一步研究,最终发现C1q敲除对脑出血损伤机制兼具利弊,但注射凝血酶后有持续益处,平衡C1q在脑出血后的作用或为潜在治疗方向[30]。"

2.6 关键词突现分析 关键词的突现功能可以直观地展示某些特定关键词在某一时段频次突然增加,可以代表在当时段的研究热点[31] 。通过对文献进行关键词突现分析,筛选得排名前10的关键词有activation,brain,differentiation,disease,analysis reveals,inhibition,microglia,system,reveals,angiogenesis (图9),可见细胞异质性、小胶质细胞激活和血管生成等机制已成为单细胞转录组测序技术在脑卒中领域研究的热门主题,提示这些细胞和相关机制可能是当前研究的前沿和热点。如2021年,GUO等[27]首次使用单细胞转录组测序技术研究缺血性脑卒中急性期小鼠皮质半暗区的细胞异质性和分子变化,提示小胶质细胞在缺血性脑卒中中的重要作用;通过 RNAscope 原位杂交和免疫荧光染色,验证了相关基因在对应细胞中的特异性表达,确保了单细胞测序结果的可靠性,该研究首次构建了缺血性脑卒中急性期半暗带细胞图谱,为干预神经炎症、促进神经修复提供了理论支撑,也为缺血性脑卒中的精准治疗提供了新方向,推动了脑卒中治疗从传统干预向精准调控的转变,还为其他神经系统疾病的单细胞研究奠定基础。单细胞转录组测序技术还为揭示血管生成与炎症的相互作用提供了关键手段,例如通过单细胞转录组测序技术发现激活紊乱血流的 JAG1-NOTCH4信号通路能够抑制内皮细胞增殖和迁移,打破内皮修复与炎症的平衡,从而驱动斑块形成,精准解析血管生成相关细胞亚群(如高表达血管内皮生长因子或促炎因子的内皮细胞)的异质性,结合功能实验(如基因敲除、抗体阻断)验证关键通路(如Notch、核因子κB)在调控血管新生与炎症微环境中的协同作用[32]。基于单细胞数据的靶点筛选(如JAG1抑制剂)和患者分层策略(如修复型内皮亚群占比),为开发靶向血管生成-炎症轴的治疗方案提供了新思路,凸显单细胞测序在动态解析疾病机制及精准干预中的核心价值。一项关于单细胞测序分析研究显示,两个特定基因Mlkl与Nlrp3通过调控内皮细胞和小胶质细胞的程序性死亡,成为连接脑卒中与重度抑郁症的关键分子桥梁,该研究为理解两种疾病的共病机制及异病同治奠定了重要基础[33]。如今科研人员越来越认识到细胞异质性在脑卒中研究中的关键作用,而单细胞转录组测序技术作为解决这一问题的有效手段,将在未来的研究中继续发挥重要作用。"

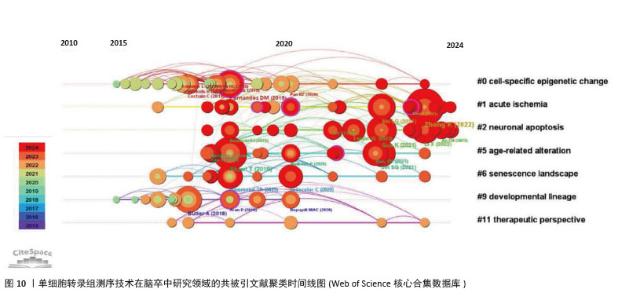
2.7 文献共被引分析 通过共被引文献聚类时间线图(图10),可以清楚地了解该领域的演化路径[34]。聚类#0细胞特异性表观遗传变化的研究表明,单细胞转录组测序技术为单细胞水平解析疾病中细胞基因表达特征、鉴定特异性细胞类型提供高通量方法[35]。尽管技术存在局限性,但在揭示急性缺血等病理状态下细胞表观遗传修饰对基因表达的调控机制、对细胞功能或凋亡的影响等方面极具价值[36],不仅深化了对缺血性疾病分子机制的理解,也为挖掘精准治疗靶点提供了重要技术支撑。聚类#1 急性缺血的研究整合多源数据,聚焦急性缺血病理机制[37]。聚类#2 神经元凋亡作为急性缺血损伤的核心特征,与缺血性疾病预后紧密关联,深入研究其在急性缺血病理进程中的潜在作用至关重要。借助单细胞转录组测序技术发现,神经炎症过程中单细胞水平(如小胶质细胞)的精确转录变化,为基于细胞亚型特异性分子的脑卒中疾病机制探索和药物发现开辟了新领域[10]。此外,研究还筛选出部分信号通路,如在小胶质细胞中cGAS或HDAC3缺失减轻了缺血再灌注诱导的神经炎症和脑损伤[38],成为改善急性缺血性疾病预后的潜在干预方向。"
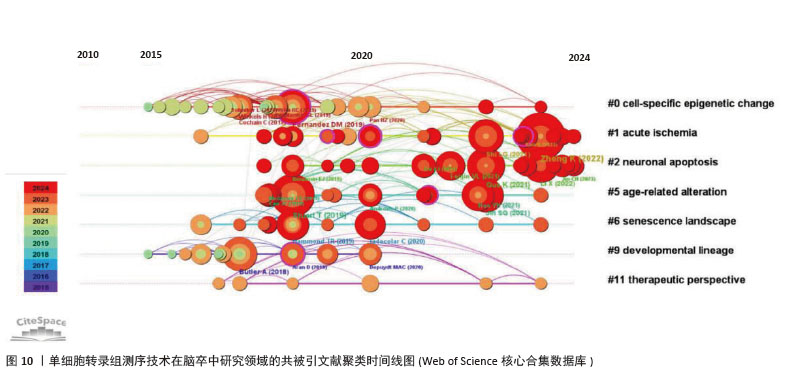
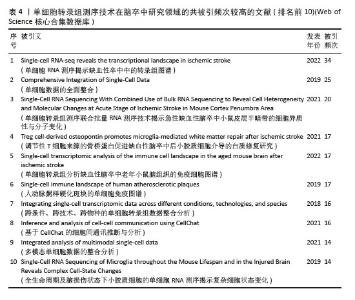
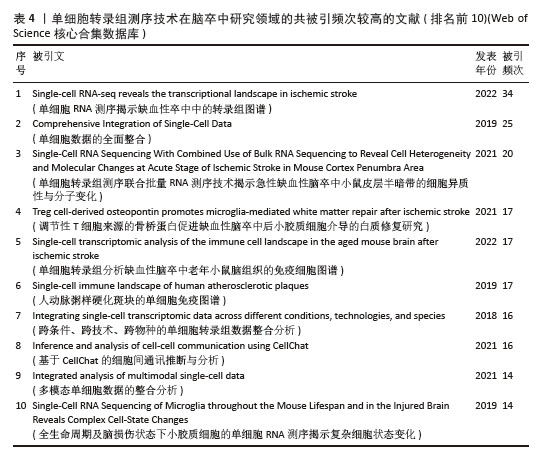
共被引频次前10篇文献,涵盖了单细胞转录组测序技术在多个方面的应用(表4),如揭示细胞异质性、免疫细胞景观、多模态单细胞数据整合、细胞间通讯等。共被引频次最高的文献“Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke”[10],主要概述了利用单细胞转录组测序技术揭示单细胞水平神经炎症的转录变化,为细胞亚型特异性分子的脑卒中疾病机制探索和药物开发开辟了新的领域。排名第二的文献“Comprehensive Integration of Single-Cell Data”报道了基于全面更新的统计模型的单细胞测序数据整合方案[39],并利用这一技术成功实现了单细胞转录组、蛋白组、表观组和空间信息数据集之间的信息迁移。这些文献的高被引频次表明它们在该领域具有重要的影响力,是后续研究的重要参考依据。科研人员可以基于这些研究成果,进一步拓展研究方向,深入探究单细胞转录组测序技术在脑卒中研究领域的应用潜力。"
| [1] HILKENS NA, CASOLLA B, LEUNG TW, et al. Stroke. Lancet. 2024;403(10446):2820-2836. [2] FAN F, YANG L, LI R, et al. Salidroside as a potential neuroprotective agent for ischemic stroke: a review of sources, pharmacokinetics, mechanism and safety. Biomed Pharmacother. 2020;129:110458. [3] REPORT ON STROKE CENTER IN CHINA WRITING GROUP.《中国卒中中心报告2022》概要[J].中国脑血管病杂志,2024, 21(8):565-576. [4] ZHAO Y, ZHANG X, CHEN X, et al. Neuronal injuries in cerebral infarction and ischemic stroke: From mechanisms to treatment (Review). Int J Mol Med. 2022;49(2):15. [5] THOMALLA G, SIMONSEN CZ, BOUTITIE F, et al. MRI-Guided Thrombolysis for Stroke with Unknown Time of Onset. N Engl J Med. 2018;379(7):611-622. [6] GHOZY S, REDA A, VARNEY J, et al. Neuroprotection in Acute Ischemic Stroke: A Battle Against the Biology of Nature. Front Neurol. 2022;13:870141. [7] XIONG Y, WAKHLOO AK, FISHER M. Advances in Acute Ischemic Stroke Therapy. Circ Res. 2022;130(8):1230-1251. [8] 汪茂林,杨洪军.单细胞转录组测序技术在药物研究中的应用[J].药学学报,2023, 58(9):2551-2559. [9] ZHENG J, RU W, ADOLACION JR, et al. Single-cell RNA-seq analysis reveals compartment-specific heterogeneity and plasticity of microglia. iScience. 2021; 24(3):102186. [10] ZHENG K, LIN L, JIANG W, et al. Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke. J Cereb Blood Flow Metab. 2022;42(1):56-73. [11] TRAN MN, MAYNARD KR, SPANGLER A, et al. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron. 2021;109(19): 3088-3103.e5. [12] TASIC B. Single cell transcriptomics in neuroscience: cell classification and beyond. Curr Opin Neurobiol. 2018;50: 242-249. [13] JOVIC D, LIANG X, ZENG H, et al. Single-cell RNA sequencing technologies and applications: A brief overview. Clin Transl Med. 2022;12(3):e694. [14] WANG H. Microglia Heterogeneity in Alzheimer’s Disease: Insights From Single-Cell Technologies. Front Synaptic Neurosci. 2021;13:773590. [15] YANG B, HU S, JIANG Y, et al. Advancements in Single-Cell RNA Sequencing Research for Neurological Diseases. Mol Neurobiol. 2024;61(11):8797-8819. [16] HE Z, CHEN Q, WANG K, et al. Single-cell transcriptomics analysis of cellular heterogeneity and immune mechanisms in neurodegenerative diseases. Eur J Neurosci. 2024;59(3):333-357. [17] FERNÁNDEZ-MOYA SM, GANESH AJ, Plass M. Neural cell diversity in the light of single-cell transcriptomics. Transcription. 2023;14(3-5):158-176. [18] HE Y, LU W, ZHOU X, et al. Unraveling Alzheimer’s disease: insights from single-cell sequencing and spatial transcriptomic. Front Neurol. 2024;15:1515981. [19] 张晓琳,王卫娣,周子凯.单细胞测序在神经精神疾病研究及未来精准诊疗中的应用[J].中国医药导刊,2022,24(3):211-218. [20] KE Y, JIAN-YUAN H, PING Z, et al. The progressive application of single-cell RNA sequencing technology in cardiovascular diseases. Biomed Pharmacother. 2022;154: 113604. [21] MARTINS-FERREIRA R, CALAFELL-SEGURA J, LEAL B, et al. The Human Microglia Atlas (HuMicA) unravels changes in disease-associated microglia subsets across neurodegenerative conditions. Nat Commun. 2025;16(1):739. [22] OLAH M, MENON V, HABIB N, et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat Commun. 2020;11(1):6129. [23] AHMADI A, GISPERT JD, NAVARRO A, et al. Single-cell Transcriptional Changes in Neurodegenerative Diseases. Neuroscience. 2021;479:192-205. [24] JEONG HW, DIÉGUEZ-HURTADO R, ARF H, et al. Single-cell transcriptomics reveals functionally specialized vascular endothelium in brain. Elife. 2022;11: e57520. [25] 陈悦,陈超美,刘则渊,等.CiteSpace知识图谱的方法论功能[J].科学学研究, 2015,33(2):242-253. [26] 宗淑萍.基于普赖斯定律和综合指数法的核心著者测评:以《中国科技期刊研究》为例[J].中国科技期刊研究,2016, 27(12):1310-1314. [27] GUO K, LUO J, FENG D, et al. Single-Cell RNA Sequencing With Combined Use of Bulk RNA Sequencing to Reveal Cell Heterogeneity and Molecular Changes at Acute Stage of Ischemic Stroke in Mouse Cortex Penumbra Area. Front Cell Dev Biol. 2021;9:624711. [28] SUN D, LUO F, FANG C, et al. Molecular mechanisms underlying the therapeutic effects of Linggui Zhugan decoction in stroke: Insights from network pharmacology and single-cell transcriptomics analysis. Medicine (Baltimore). 2024;103(13):e37482. [29] WANG Z, WU X, YAN T, et al. Elevated Plasma Complement C1q Levels Contribute to a Poor Prognosis After Acute Primary Intracerebral Hemorrhage: A Prospective Cohort Study. Front Immunol. 2022;13:920754. [30] FU X, YE F, WAN Y, et al. The Role of Complement C1qa in Experimental Intracerebral Hemorrhage. Transl Stroke Res. 2025;16(4):1229-1240. [31] 张勇.我国高校阅读推广研究演进路径、热点与趋势分析[J].图书馆工作与研究, 2020(8):87-97. [32] SOUILHOL C, TARDAJOS AYLLON B, LI X, et al. JAG1-NOTCH4 mechanosensing drives atherosclerosis. Sci Adv. 2022;8(35): eabo7958. [33] ZHAO Y, SUN W, FAN Q, et al. Exploring the potential molecular intersection of stroke and major depression disorder. Biochem Biophys Res Commun. 2024; 720:150079. [34] 唐荣,魏欣,马江,等.基于CiteSpace科学知识图谱分析石榴皮研究现状及热点[J].中草药,2023,54(12):3949-3961. [35] ZUCHA D, ABAFFY P, KIRDAJOVA D, et al. Spatiotemporal transcriptomic map of glial cell response in a mouse model of acute brain ischemia. Proc Natl Acad Sci U S A. 2024;121(46):e2404203121. [36] NEAL M, RICHARDSON JR. Epigenetic regulation of astrocyte function in neuroinflammation and neurodegeneration. Biochim Biophys Acta Mol Basis Dis. 2018; 1864(2):432-443. [37] KHOSHNAM SE, MOALEMNIA A, ANBIYAEE O, et al. LncRNA MALAT1 and Ischemic Stroke: Pathogenesis and Opportunities. Mol Neurobiol. 2024;61(7):4369-4380. [38] LIAO Y, CHENG J, KONG X, et al. HDAC3 inhibition ameliorates ischemia/reperfusion-induced brain injury by regulating the microglial cGAS-STING pathway. Theranostics. 2020;10(21):9644-9662. [39] STUART T, BUTLER A, HOFFMAN P, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888-1902.e21. [40] XU S, LU J, SHAO A, et al. Glial Cells: Role of the Immune Response in Ischemic Stroke. Front Immunol. 2020;11:294. [41] OCHOCKA N, KAMINSKA B. Microglia Diversity in Healthy and Diseased Brain: Insights from Single-Cell Omics. Int J Mol Sci. 2021;22(6):3027. [42] 高伟铭. 基于单细胞及空间转录组测序的放射性脑损伤研究[D].石家庄:河北大学,2023. [43] 胡静涵,胡懿凡,蒋俊锋,等.单细胞转录组测序技术及其在肿瘤研究中的应用进展[J].海军军医大学学报,2023, 44(7):800-807. [44] JI C, SHENG L, HAN K, et al. Microglial intervention in ischemic stroke: Roles and intervention strategies. Neural Regen Res. 2026;21(2):443-454. [45] KANG R, GAMDZYK M, LENAHAN C, et al. The Dual Role of Microglia in Blood-Brain Barrier Dysfunction after Stroke. Curr Neuropharmacol. 2020;18(12):1237-1249. [46] BAIK SH, KANG S, LEE W, et al. A Breakdown in Metabolic Reprogramming Causes Microglia Dysfunction in Alzheimer’s Disease. Cell Metab. 2019; 30(3):493-507.e6. [47] KALININA TS, SHISHKINA GT, LANSHAKOV DA, et al. Comparative Investigation of Expression of Glutamatergic and GABAergic Genes in the Rat Hippocampus after Focal Brain Ischemia and Central LPS Administration. Biochemistry (Mosc). 2023; 88(4):539-550. [48] LIDDELOW SA, GUTTENPLAN KA, CLARKE LE, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481-487. [49] ATTA AA, IBRAHIM WW, MOHAMED AF, et al. Microglia polarization in nociplastic pain: mechanisms and perspectives. Inflammopharmacology. 2023;31(3): 1053-1067. [50] REN J, CHE Y, LI H, et al. SGK3 deficiency in macrophages suppresses angiotensin II-induced cardiac remodeling via regulating Ndufa13-mediated mitochondrial oxidative stress. Cell Mol Life Sci. 2024; 81(1):359. [51] LIU B, SUN X, SUYEOKA G, et al. TGFβ signaling induces expression of Gadd45b in retinal ganglion cells. Invest Ophthalmol Vis Sci. 2013;54(2):1061-1069. [52] WU DD, ZHANG F, HAO F, et al. Matricellular protein Cyr61 bridges lysophosphatidic acid and integrin pathways leading to cell migration. J Biol Chem. 2014;289(9): 5774-5783. [53] LUO L, GRIBSKOV M, WANG S. Bibliometric review of ATAC-Seq and its application in gene expression. Brief Bioinform. 2022; 23(3):bbac061. |
| [1] | Zhu Xiaolong, Zhang Wei, Yang Yang. Visualization analysis of research hotspots and cutting-edge information in the field of intervertebral disc regeneration and repair [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(9): 2391-2402. |
| [2] | Wang Zheng, Cheng Ji, Yu Jinlong, Liu Wenhong, Wang Zhaohong, Zhou Luxing. Progress and future perspectives on the application of hydrogel materials in stroke therapy [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(8): 2081-2090. |
| [3] | Gao Feng, Zhang Jun, Yu Wenjun, Chanyu Yujing, Zhao Le, Hu Yuting, Wang Junhua, Liu Yongfu. Effects of wrist-hand orthosis on hand dysfunction in stroke patients: a meta-analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(8): 2124-2131. |
| [4] | Lai Yu, Chen Yueping, Zhang Xiaoyun. Research hotspots and frontier trends of bioactive materials in treating bone infections [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(8): 2132-2144. |
| [5] | Cai Ziming, Yu Qinghe, Ma Pengfei, Zhang Xin, Zhou Longqian, Zhang Chongyang, Lin Wenping. Heme oxygenase-1 alleviates lipopolysaccharide-induced inflammatory response in nucleus pulposus mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1624-1631. |
| [6] | He Jiale, Huang Xi, Dong Hongfei, Chen Lang, Zhong Fangyu, Li Xianhui. Acellular dermal matrix combined with adipose-derived stem cell exosomes promotes burn wound healing [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1699-1710. |
| [7] | Xia Linfeng, Wang Lu, Long Qianfa, Tang Rongwu, Luo Haodong, Tang Yi, Zhong Jun, Liu Yang. Human umbilical cord mesenchymal stem cell-derived exosomes alleviate blood-brain barrier damage in mice with septic encephalopathy [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1711-1719. |
| [8] | Cui Lianxu, Li Haomin, Xu Junrong, Tan Baodong, Lu Dahong, Peng Siwei, Wang Jinhui. Effect of umbilical cord mesenchymal stem cell conditioned medium on tissue repair after traumatic craniocerebral injury in miniature pigs [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1730-1735. |
| [9] | Tao Daiju, Su Haiyu, Wang Yuqi, Shen Zhiqiang, He Bo . Construction and identification of stable PC12 cell lines with high/low expression of miR-122-5p [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1790-1799. |
| [10] | Cao Yong, Teng Hongliang, Tai Pengfei, Li Junda, Zhu Tengqi, Li Zhaojin. Interactions between cytokines and satellite cells in muscle regeneration [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1808-1817. |
| [11] | Hou Chaowen, Li Zhaojin, Kong Jianda, Zhang Shuli. Main physiological changes in skeletal muscle aging and the multimechanism regulatory role of exercise [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1464-1475. |
| [12] | You Huijuan, Wu Shuzhen, Rong Rong, Chen Liyuan, Zhao Yuqing, Wang Qinglu, Ou Xiaowei, Yang Fengying. Macrophage autophagy in lung diseases: two-sided effects [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1516-1526. |
| [13] | Guo Ying, Tian Feng, Wang Chunfang. Potential drug targets for the treatment of rheumatoid arthritis: large sample analysis from European databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1549-1557. |
| [14] | Yin Yongcheng, Zhao Xiangrui, Yang Zhijie, Li Zheng, Li Fang, Ning Bin. Effect and mechanism of peroxiredoxin 1 in microglial inflammation after spinal cord injury [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1106-1113. |
| [15] | Zhang Di, Zhao Jun, Ma Guangyue, Sun Hui, Jiang Rong. Mechanism of depression-like behavior in chronic social defeat stress mice based on high-throughput sequencing [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1139-1146. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||