中国组织工程研究 ›› 2016, Vol. 20 ›› Issue (7): 947-956.doi: 10.3969/j.issn.2095-4344.2016.07.005
• 组织构建细胞学实验 cytology experiments in tissue construction • 上一篇 下一篇
人基质金属蛋白酶3基因RNA慢病毒载体构建及在人退变髓核细胞中的鉴定
曹 进,傅培荣,房 晶,杨建坤,位华卫,李思源,高 峰,西永明
- 青岛大学附属医院脊柱外科,山东省青岛市 266003
-
收稿日期:2015-11-24出版日期:2016-02-12发布日期:2016-02-12 -
通讯作者:西永明,博士,副教授,主任医师,青岛大学附属医院脊柱外科,山东省青岛市 266000 -
作者简介:曹进,男,1985年生,河南省周口市人,汉族,青岛大学在读硕士,主要从事骨科的基础和临床研究。 -
基金资助:国家自然科学基金(81470104)
Constructing and identifying a lentiviral vector of RNA interference targeting matrix metalloproteinases-3 gene in human degenerative nucleus pulposus cells
- Department of Spine Surgery, Affiliated Hospital of Qingdao University, Qingdao 266003, Shandong Province, China
-
Received:2015-11-24Online:2016-02-12Published:2016-02-12 -
Contact:Xi Yong-ming, M.D., Associate professor, Chief physician, Department of Spine Surgery, Affiliated Hospital of Qingdao University, Qingdao 266003, Shandong Province, China -
About author:Cao Jin, Studying for master’s degree, Department of Spine Surgery, Affiliated Hospital of Qingdao University, Qingdao 266003, Shandong Province, China -
Supported by:the National Natural Science Foundation of China, No. 81470104
摘要:
文章快速阅读:
.jpg)
文题释义:
髓核:是位于软骨板和纤维环中间,由纵横交错的纤维网状结构即软骨细胞和蛋白多糖黏液样基质构成的弹性胶冻物质。髓核在出生时体积大而松散,位于椎间盘的中央,至成年时位置移至椎间盘的中后部。在20岁以前构成髓核的主要物质是大量蛋白多糖复合体、胶原纤维和纤维软骨,随着年龄的增长,髓核中的蛋白多糖解聚增多,水分逐渐减少,胶原增粗并逐渐被纤维软骨所替代。
ORCID:0000-0002-2921-0699(Xi Yong-ming)
引用本文
曹 进,傅培荣,房 晶,杨建坤,位华卫,李思源,高 峰,西永明. 人基质金属蛋白酶3基因RNA慢病毒载体构建及在人退变髓核细胞中的鉴定[J]. 中国组织工程研究, 2016, 20(7): 947-956.
Cao Jin, Fu Pei-rong, Fang Jing, Yang Jian-kun, Wei Hua-wei, Li Si-yuan, Gao Feng, Xi Yong-ming. Constructing and identifying a lentiviral vector of RNA interference targeting matrix metalloproteinases-3 gene in human degenerative nucleus pulposus cells[J]. Chinese Journal of Tissue Engineering Research, 2016, 20(7): 947-956.
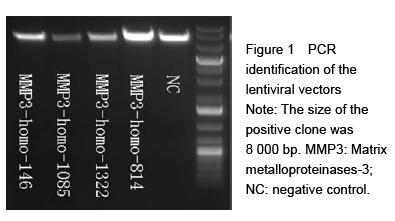
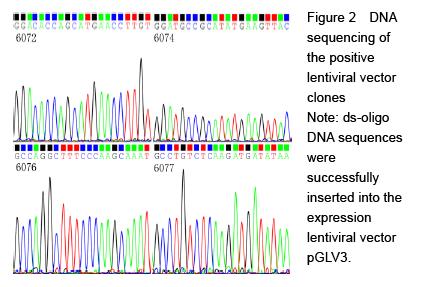
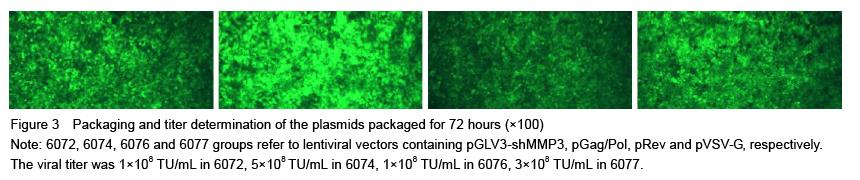
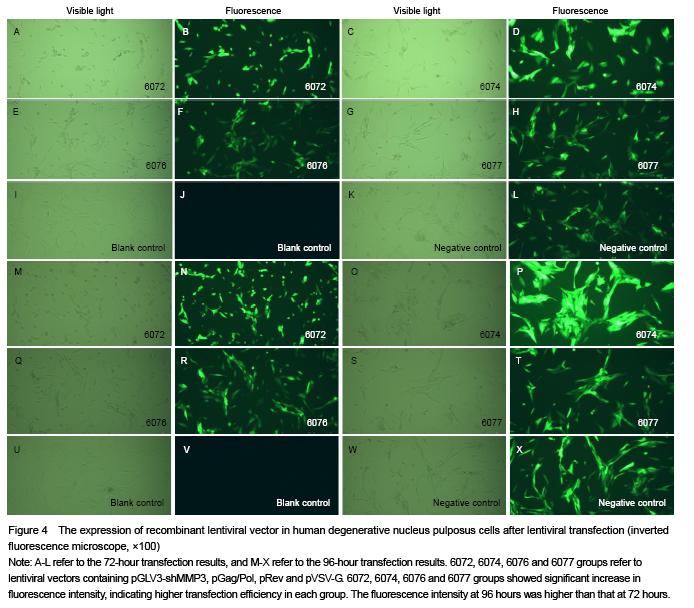
.jpg)
| [1] Kadow T, Sowa G, Vo N, et al. Molecular basis of intervertebral disc degeneration and herniations: what are the important translational questions? Clin Orthop Relat Res. 2015;473(6):1903-1912. [2] Hoy D, Bain C, Williams G, et al. A systematic review of the global prevalence of low back pain. Arthritis Rheum. 2012;64(6):2028-2037. [3] Weber KT, Jacobsen TD, Maidhof R, et al. Developments in intervertebral disc disease research: pathophysiology, mechanobiology, and therapeutics. Curr Rev Musculoskelet Med. 2015;8(1):18-31. [4] Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10(1):44-56. [5] Roberts S, Caterson B, Menage J, et al. Matrix metalloproteinases and aggrecanase: their role in disorders of the human intervertebral disc. Spine (Phila Pa 1976). 2000;25(23):3005-3013. [6] Weiler C, Nerlich AG, Zipperer J, et al. 2002 SSE Award Competition in Basic Science: expression of major matrix metalloproteinases is associated with intervertebral disc degradation and resorption. Eur Spine J. 2002;11(4):308-320. [7] Le Maitre CL, Freemont AJ, Hoyland JA. Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J Pathol. 2004; 204(1):47-54. [8] Wang X, Li F, Fan C, et al. Effects and relationship of ERK1 and ERK2 in interleukin-1β-induced alterations in MMP3, MMP13, type II collagen and aggrecan expression in human chondrocytes. Int J Mol Med. 2011;27(4):583-589. [9] Murphy G, Cockett MI, Stephens PE, et al. Stromelysin is an activator of procollagenase. A study with natural and recombinant enzymes. Biochem J. 1987;248(1): 265-268. [10] Bachmeier BE, Nerlich A, Mittermaier N, et al. Matrix metalloproteinase expression levels suggest distinct enzyme roles during lumbar disc herniation and degeneration. Eur Spine J. 2009;18(11):1573-1586. [11] Yurube T, Nishida K, Suzuki T, et al. Matrix metalloproteinase (MMP)-3 gene up-regulation in a rat tail compression loading-induced disc degeneration model. J Orthop Res. 2010;28(8):1026-1032. [12] Yurube T, Takada T, Suzuki T, et al. Rat tail static compression model mimics extracellular matrix metabolic imbalances of matrix metalloproteinases, aggrecanases, and tissue inhibitors of metalloproteinases in intervertebral disc degeneration. Arthritis Res Ther. 2012;14(2):R51. [13] Canbay S, Turhan N, Bozkurt M, et al. Correlation of matrix metalloproteinase-3 expression with patient age, magnetic resonance imaging and histopathological grade in lumbar disc degeneration. Turk Neurosurg. 2013;23(4):427-433. [14] Wei F, Zhong R, Zhou Z, et al. In vivo experimental intervertebral disc degeneration induced by bleomycin in the rhesus monkey. BMC Musculoskelet Disord. 2014;15:340. [15] Jing W, Jiang W. MicroRNA-93 regulates collagen loss by targeting MMP3 in human nucleus pulposus cells. Cell Prolif. 2015;48(3):284-292. [16] Mocellin S, Costa R, Nitti D. RNA interference: ready to silence cancer? J Mol Med (Berl). 2006;84(1):4-15. [17] Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006): 343-349. [18] McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3(10): 737-747. [19] Gencer S, Cebeci A, Irmak-Yazicioglu MB. Silencing of the MMP3 gene by siRNA transfection in gastric cancer AGS cells. J Gastrointestin Liver Dis. 2011;20(1):19-26. [20] Hegedüs L, Cho H, Xie X, et al. Additional MDA-MB-231 breast cancer cell matrix metalloproteinases promote invasiveness. J Cell Physiol. 2008;216(2):480-485. [21] Pfirrmann CW, Metzdorf A, Zanetti M, et al. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976). 2001;26(17): 1873-1878. [22] Nishida K, Suzuki T, Kakutani K, et al. Gene therapy approach for disc degeneration and associated spinal disorders. Eur Spine J. 2008;17 Suppl 4:459-466. [23] Dyawanapelly S, Ghodke SB, Vishwanathan R, et al. RNA interference-based therapeutics: molecular platforms for infectious diseases. J Biomed Nanotechnol. 2014;10(9):1998-2037. [24] Baigude H, Rana TM. Strategies to antagonize miRNA functions in vitro and in vivo. Nanomedicine (Lond). 2014;9(16):2545-2555. [25] Ren XF, Diao ZZ, Xi YM, et al. Adeno-associated virus-mediated BMP-7 and SOX9 in vitro co-transfection of human degenerative intervertebral disc cells. Genet Mol Res. 2015;14(2):3736-3744. [26] Sakuma T, Barry MA, Ikeda Y. Lentiviral vectors: basic to translational. Biochem J. 2012;443(3):603-618. [27] Pluta K, Kacprzak MM. Use of HIV as a gene transfer vector. Acta Biochim Pol. 2009;56(4):531-595. [28] Wu J, Wang D, Ruan D, et al. Prolonged expansion of human nucleus pulposus cells expressing human telomerase reverse transcriptase mediated by lentiviral vector. J Orthop Res. 2014;32(1):159-166. [29] Ma X, Lin Y, Yang K, et al. Effect of lentivirus-mediated survivin transfection on the morphology and apoptosis of nucleus pulposus cells derived from degenerative human disc in vitro. Int J Mol Med. 2015;36(1):186-194. |
| No related articles found! |
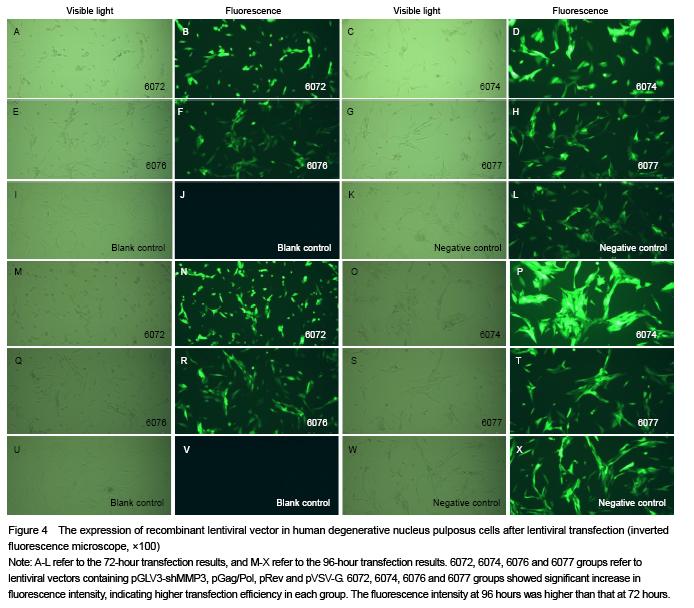
As shown in Table 1, four pairs of MMP3 shRNA were designed with Designer3.0 software (GenePharma, Shanghai, China) according to the human MMP3 mRNPA (NM_002422.4) sequence obtained from GenBank (http://www.ncbi.nlm.nih. gov/genbank) and the shRNA design principles. Double-stranded DNA fragments, with cohesive termini of the EcoRI and BamHI restriction enzymes (MBI Fermentas) and hairpin sequence, were synthesized in vitro.
.jpg)
Annealing of DNA oligo sequences targeting MMP3 gene
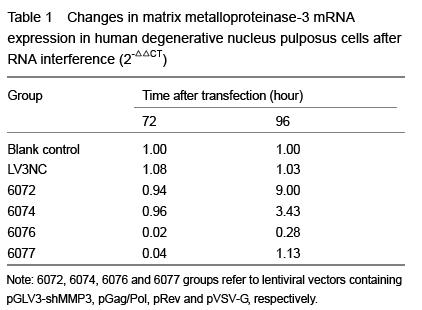
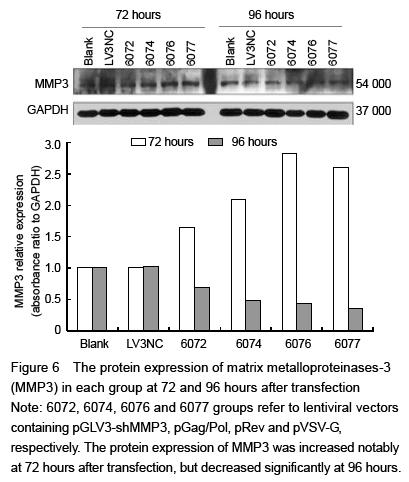
At 72 and 96 hours after transfection, cells were fully cracked by Trizol reagent, and chloroform was added into the supernatants. Blended by concussion, the supernatants were added isopropyl alcohol and centrifuged, and then the mRNA was diluted with ultrapure water after being washed with 75% ethanol. Absorbance (A) value and concentration of the RNA were measured with the spectrophotometer, and the purity was detected by RNA electrophoresis. Total RNA was extracted and cDNA was synthesized with RT-PCR following the reaction conditions: 42 ℃ 30 minutes, 85 ℃ 10 minutes. The RT reaction conditions: 2×RT buffer 10 μL, RT primer (1 μmol/L) 1.2 μL, total RNA 2 μL, MMLV reverse transcriptase (200 U/μL) 0.2 μL, DEPC water 6.6 μL, total 20 μL. Then real-time fluorescence quantitative PCR was carried out with RTFQ PCR kit following the instruction. The reaction conditions were as follows: pre-denaturation at 95 ℃ for 3 minutes, denaturation at 95 ℃ for 12 seconds, annealing at 62 ℃ for 40 seconds, total 40 cycles. Then the mixtures were denatured at 95 ℃ for 60 seconds at the end of the PCR process, and cooled to 55 ℃. Sequences of the primer refer to Table 2. Reaction products were examined with 1% agarose gel electrophoresis. A values of the electrophoresis bands were analyzed with gel imaging system to verify the stripes of target gene. Quantitative analysis was performed using the ratio of the target gene to hACTB. The PCR reaction conditions: 2× quantitative PCR Master Mix 10.0 μL, primer F (20 μmol/L) 0.08 μL, primer R (20 μmol/L) 0.08 μL, cDNA 2 μL, Taq DNA polymerase (2.5 U/μL) 0.4 μL, ddH2O 7.44 μL, total 20 μL.
.jpg)
文章快速阅读:
.jpg)
文题释义:
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||