Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (22): 5886-5896.doi: 10.12307/2026.212
Systematic druggable genome-wide Mendelian randomization identifies therapeutic targets for major depressive disorder
Zhou Menghan1, Liu Shuning2, Jiang Tao3, Sun Zhuangzhuang1, Cao Lingling1, Su Xin3, Yu Cheng1, Guo Junpeng1
- 1College of Clinical Medicine, Changchun University of Chinese Medicine, Changchun 130117, Jilin Province, China; 2School of Marxism, Changchun University of Chinese Medicine, Changchun 130117, Jilin Province, China; 3College of Basic Medicine, Changchun University of Chinese Medicine, Changchun 130117, Jilin Province, China
-
Received:2025-08-26Accepted:2025-09-07Online:2026-08-08Published:2025-12-29 -
Contact:Guo Junpeng, Professor, Doctoral supervisor, College of Clinical Medicine, Changchun University of Chinese Medicine, Changchun 130117, China Co-corresponding author: Yu Cheng, Associate professor, College of Clinical Medicine, Changchun University of Chinese Medicine, Changchun 130117, Jilin Province, China -
About author:Zhou Menghan, MS, College of Clinical Medicine, Changchun University of Chinese Medicine, Changchun 130117, Jilin Province, China -
Supported by:Natural Science Foundation of Jilin Province, No. YDZJ202501ZYTS192 (to CLL)
CLC Number:
Cite this article
Zhou Menghan, Liu Shuning, Jiang Tao, Sun Zhuangzhuang, Cao Lingling, Su Xin, Yu Cheng, Guo Junpeng. Systematic druggable genome-wide Mendelian randomization identifies therapeutic targets for major depressive disorder[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(22): 5886-5896.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
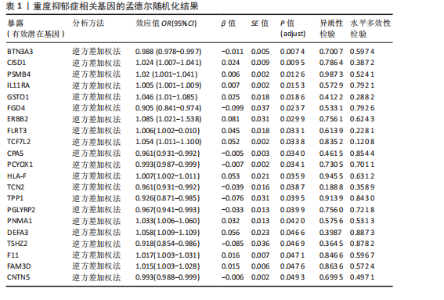
2.1 候选药物相关基因的识别 从DGIdb数据库共检索到2 725个可成药基因,通过系统性文献综述额外识别出4 302个可成药基因,合并数据集并去除重复项后,整理出包含6 959个不重叠的可成药基因的综合列表用于后续分析。pQTL数据来源于UKB-PPP和deCODE Genetics,用于提取血浆蛋白质遗传关联的汇总统计数据,合并数据集并去除重复项后获得10 001个pQTL整合数据。UKB-PPP覆盖了54 219名UK Biobank参与者队列中2 923种蛋白质的血浆pQTL数据。deCODE Genetics则包含了从4 907名冰岛人进行的35 559次适体检测中所获得的7 258种血浆蛋白水平数据。设定错误发现率(False Discovery Rate,FDR) < 0.05作为显著性阈值。这些数据集的详细描述可在原始文献的补充表格中找到。 2.2 重度抑郁症有效基因的筛选 利用eQTLGen联盟提供的eQTL数据和pQTL数据对筛选出的6 959个和10 001个可成药基因进行了潜在暴露变量的评估。在eQTL数据集和pQTL数据集中分别鉴定出2 478个和7 954个可成药基因,对这些基因进行孟德尔随机化分析。采用Benjamini-Hochberg FDR法校正P值,识别出21个与重度抑郁症显著相关的可成药基因,其中BTN3A3(嗜乳脂蛋白亚家族3成员A3)、CISD1(CDGSH铁硫结构域1)和PSMB4(蛋白酶体20S亚基β4)最值得关注(表1)。对鉴定出的基因进行进一步的敏感性分析以评估多效性和异质性,未观察到显著的多效性效应或异质性证据。 2.3 定位分析结果 在21个显著基因中,结果显示BTN3A3(H4.abf=0.642)、PSMB4(H4.abf=0.574)和 CISD1(H4.abf=0.522)的H4.abf 值均超过0.5,表明通过孟德尔随机化分析鉴定出的BTN3A3、PSMB4、CISD1与重度抑郁症之间高度存在共享因果变异的"
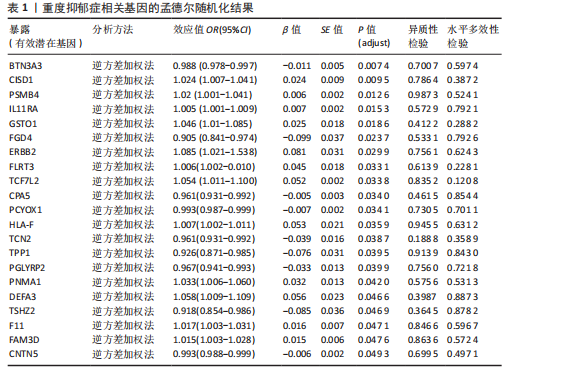
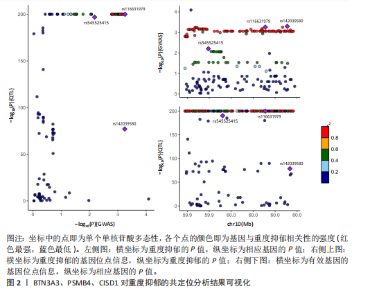
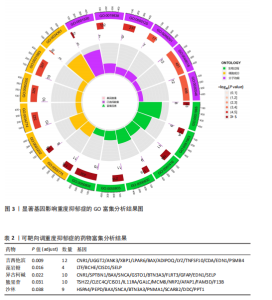
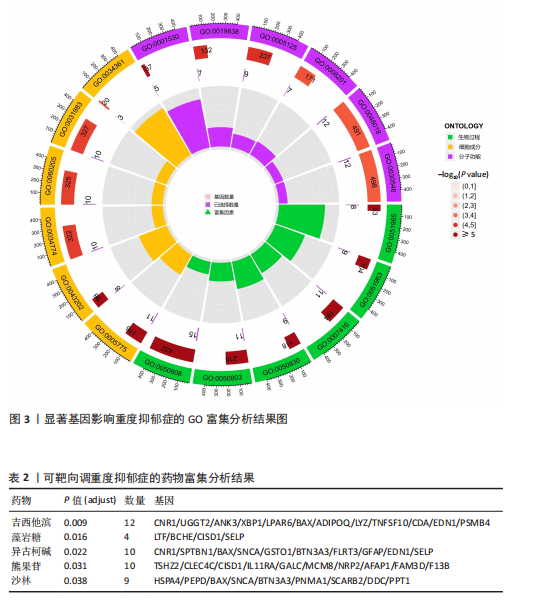
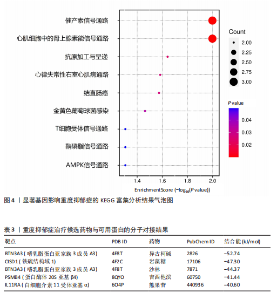
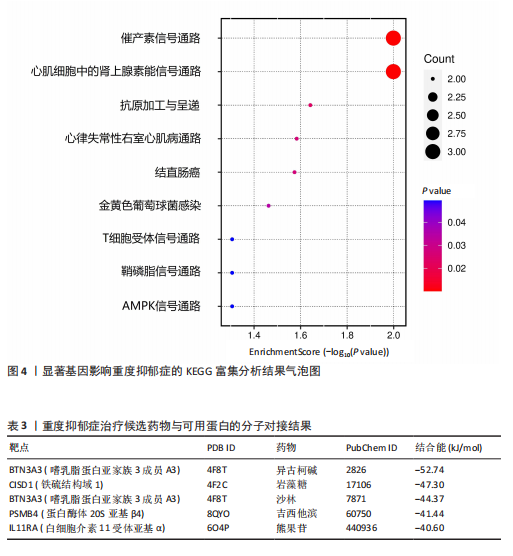
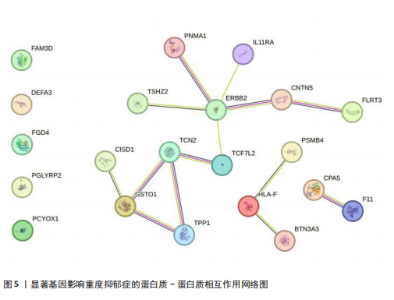
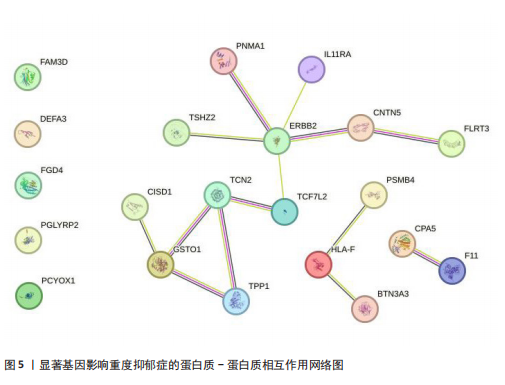
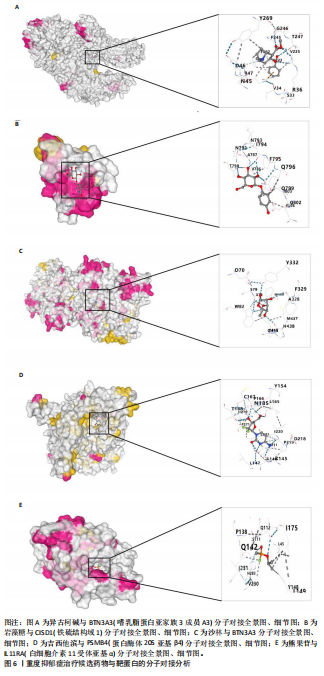
可能性(图2)。 2.4 显著基因功能分析 对21个潜在靶点进行GO富集分析,可以看出这些候选基因主要涉及“抗原加工与呈递”“蛋白质降解与加工”“线粒体外膜”“免疫受体活性”等多个与重度抑郁相关的功能通路(图3)。具体而言,生物过程中,“抗原加工与呈递”和“蛋白质水解过程”显著富集,提示候选基因可能调控重度抑郁症患者的免疫激活状态与细胞应激反应,在抗原呈递过程在该过程中可能发挥促进作用[35];细胞成分中,多个候选基因富集于“线粒体外膜”与“蛋白酶体复合体”,线粒体功能异常被认为是抑郁症的重要发病机制之一,尤其在中枢神经系统中能量代谢不稳可影响神经递质合成与神经元可塑性[36],说明它们可能参与调控细胞能量代谢和蛋白质稳态,与慢性应激诱导下的能量障碍、线粒体功能紊乱及突触结构异常密切相关;分子功能中,富集项如“免疫受体活性”与“结合酶调节活性”提示候选基因可能通过调节免疫通路信号的识别与转导,免疫受体在细胞识别炎症信号并介导跨膜信息传导中扮演关键角色,说明它们可能与慢性疼痛或慢性低度炎症密切相关[37]。 为深入探究这些可成药基因在抑郁症中潜在的治疗机制,进一步进行了KEGG通路分析。结果显示,重度抑郁相关的显著可成药基因主要集中于免疫调控、代谢应答和信号转导相关的关键通路。例如,“抗原加工与呈递”和“T细胞受体信号通路”均与重度抑郁症中常见的免疫激活及外周炎症反应密切相关。慢性应激状态下,外周免疫系统活化和T细胞功能失衡可通过炎症因子影响中枢神经系统功能,导致情绪调节失衡。“鞘磷脂信号通路”和“AMP依赖的蛋白激酶信号通路”与能量代谢和细胞存活密切相关。研究表明,鞘磷脂代谢失衡与神经元膜结构损伤和凋亡有关,而AMP依赖的蛋白激酶作为能量代谢调控的核心分子,可影响神经元可塑性、突触传递和应激反应,在抑郁症模型中已被证实是潜在的抗抑郁药物靶点[38]。另外,其他通路如“催产素信号”则直接与情绪调控、社会行为和压力反应相关,催产素水平下降已在多项抑郁症临床研究中观察到[39],提示该调节通路可能在重度抑郁治疗中具有潜力(图4)。 2.5 靶蛋白的蛋白质-蛋白质相互作用网络 利用STRING数据库构建了包含这 21个药物靶基因的蛋白质-蛋白质相互作用网络,如图5所示。该网络包含代表蛋白质的 21个节点和代表相互作用的14条边,网络的局部聚类系数为 0.508,表明所鉴定蛋白间存在中等程度的连通性。 BTN3A3、PSMB4与CISD1在网络中具有较多的直接互作伙伴,处于互作网络的中心区域,提示这些蛋白可能为调控抑郁症关键路径的“中枢节点”。其中,PSMB4作为蛋白酶体20S核心粒子的组成部分,广泛参与细胞内蛋白降解,与神经免疫调节密切相关[40]。CISD1则与线粒体铁代谢调控有关,在调节神经元氧化应激反应中可能发挥重要作用[41]。这些关键蛋白在网络中的突出位置,提示其可能在重度抑郁的发病机制中处于核心调控地位,为靶向干预提供了理论依据。 2.6 药物富集结果 利用药物特征基因集数据库鉴定潜在的治疗化合物,根据关联基因的数量筛选出前5位候选药物作为潜在的治疗药物(表2)。值得注意的是,吉西他滨(CID 60750)、岩藻糖(CID 17106)和异古柯碱(CID 2826)的结果最为突出,它们分别与BTN3A3、PSMB4和CISD1相关联,并与大多数已鉴定的可成药基因存在联系,表明它们具有调节重度抑郁症相关生物学通路的潜力。 2.7 候选药物的分子对接分析 分子对接的结合能计算揭示了5个稳定的药物-蛋白质相互作用(表3),每次相互作用产生结合能,结合能越低,结合效果越好、亲和力越高。对接结果的可视化分别为结合全景图与局部细节图,突出了参与结合的关键氨基酸残基以及氢键长度(图6)。其中,BTN3A3表现出最低的结合能(-52.74 kJ/mol),表明它具有高度稳定的结合亲和力及潜在的治疗相关性。在BTN3A3-异古柯碱对接模型中,异古柯碱通过2个氢键与残基Ser76和Glu112形成稳定连接,同时形成疏水作用网络覆盖Val80与Ile115,有助于稳定其构象[42];在CISD1-岩藻糖对接模型中,通过极性相互作用与残基Asn39和Tyr42形成稳定键合,提示该位点可能为药物敏感结构域[43];在PSMB4-吉西他滨对接模型中,吉西他滨通过氢键与Thr20和Gln26结"
| [1] SOLIS EC, CARLIER IVE, KAMMINGA N, et al. Economic Evaluation of Self-Management for Patients with Persistent Depressive Disorder and their Caregivers. J Ment Health Policy Econ. 2024;27(4): 129-143. [2] ZHANG ZQ, YANG MH, GUO ZP, et al. Increased prefrontal cortex connectivity associated with depression vulnerability and relapse. J Affect Disord. 2022;304:133-141. [3] KESHAVARZ K, HEDAYATI A, REZAEI M, et al. Economic burden of major depressive disorder: a case study in Southern Iran. BMC Psychiatry. 2022; 22(1):577. [4] ANDERSON E, CRAWFORD CM, FAVA M, et al. Depression - Understanding, Identifying, and Diagnosing. N Engl J Med. 2024;390(17):e41. [5] ŁYSIK A, LOGOŃ K, SZCZYGIŁ A, et al. Innovative approaches in the treatment-resistant depression: exploring different therapeutic pathways. Geroscience. 2025. doi: 10.1007/s11357-025-01615-8. [6] RAKEL RE. Depression. Prim Care. 1999; 26(2):211-224. [7] FERKINGSTAD E, SULEM P, ATLASON BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021;53(12):1712-1721. [8] FRESHOUR SL, KIWALA S, COTTO KC, et al. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144-D1151. [9] QIU-QIANG Z, WEI-WEI Y, SHAN-SHU H, et al. Mendelian randomization of individual sleep traits associated with major depressive disorder. J Affect Disord. 2024;365:105-111. [10] LI Y, LV MR, WEI YJ, et al. Dietary patterns and depression risk: A meta-analysis. Psychiatry Res. 2017;253:373-382. [11] MENG X, NAVOLY G, GIANNAKOPOULOU O, et al. Multi-ancestry genome-wide association study of major depression aids locus discovery, fine mapping, gene prioritization and causal inference. Nat Genet. 2024;56(2):222-233. [12] JAMET C, DUBERTRET C, LE STRAT Y, et al. Age of onset of major depressive episode and association with lifetime psychiatric disorders, health-related quality of life and impact of gender: A cross sectional and retrospective cohort study. J Affect Disord. 2024;363:300-309. [13] BRUFFAERTS R, CAYWOOD K, AXINN WG. Early-life risk factors for depression among young adults in the United States general population: Attributable risks and gender differences. J Affect Disord. 2024;363:206-213. [14] LIU S, XU D. Causal relationship between educational attainment and chronic pain: A Mendelian randomization study. Medicine (Baltimore). 2024;103(37): e39301. [15] WANG Z, LI S, CAI G, et al. Mendelian randomization analysis identifies druggable genes and drugs repurposing for chronic obstructive pulmonary disease. Front Cell Infect Microbiol. 2024; 14:1386506. [16] VÖSA U, CLARINGBOULD A, WESTRA HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021; 53(9):1300-1310. [17] FINAN C, GAULTON A, KRUGER FA, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017; 9(383):eaag1166. [18] SUN BB, CHIOU J, TRAYLOR M, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023; 622(7982):329-338. [19] KELLY KM, SMITH JA, MEZUK B. Depression and interleukin-6 signaling: A Mendelian Randomization study. Brain Behav Immun. 2021;95:106-114. [20] ZHOU H, JI Y, SUN L, et al. Exploring the causal relationships and mediating factors between depression, anxiety, panic, and atrial fibrillation: A multivariable Mendelian randomization study. J Affect Disord. 2024; 349:635-645. [21] WANG Z, LIN X, CHEN X, et al. Genetic causality and metabolite pathway identifying the relationship of blood metabolites and psoriasis. Skin Res Technol. 2024;30(7):e13840. [22] FAN M, YUN Z, YUAN J, et al. Genetic insights into therapeutic targets for gout: evidence from a multi-omics mendelian randomization study. Hereditas. 2024;161(1):56. [23] STALEY JR, BLACKSHAW J, KAMAT MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016; 32(20):3207-3209. [24] BURGESS S, DAVEY SMITH G, DAVIES NM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 2023;4:186. [25] ZHANG C, HE Y, LIU L. Identifying therapeutic target genes for migraine by systematic druggable genome-wide Mendelian randomization. J Headache Pain. 2024;25(1):100. [26] GIAMBARTOLOMEI C, VUKCEVIC D, SCHADT EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014; 10(5):e1004383. [27] RASOOLY D, PELOSO GM, GIAMBARTOLOMEI C. Bayesian Genetic Colocalization Test of Two Traits Using coloc. Curr Protoc. 2022; 2(12):e627. [28] GIAMBARTOLOMEI C, ZHENLI LIU J, ZHANG W, et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018; 34(15):2538-2545. [29] IMPRIALOU M, PETRETTO E, BOTTOLO L. Expression QTLs Mapping and Analysis: A Bayesian Perspective. Methods Mol Biol. 2017;1488:189-215. [30] HU S, LUO Y, ZHANG Z, et al. Protein function annotation based on heterogeneous biological networks. BMC Bioinformatics. 2022;23(1):493. [31] SZKLARCZYK D, KIRSCH R, KOUTROULI M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023; 51(D1):D638-D646. [32] KIM S, CHEN J, CHENG T, et al. PubChem 2023 update. Nucleic Acids Res. 2023; 51(D1):D1373-D1380. [33] BERMAN HM, WESTBROOK J, FENG Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235-242. [34] MORRIS GM, HUEY R, LINDSTROM W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785-2791. [35] WU H, HUANG J, ZHONG Y, et al. DrugSig: A resource for computational drug repositioning utilizing gene expression signatures. PLoS One. 2017; 12(5):e0177743. [36] BHATTACHARYYA U, JOHN J, LAM M, et al. Circulating Blood-Based Proteins in Psychopathology and Cognition: A Mendelian Randomization Study. JAMA Psychiatry. 2025;82(5):481-491. [37] TKACHEV A, STEKOLSHCHIKOVA E, VANYUSHKINA A, et al. Lipid Alteration Signature in the Blood Plasma of Individuals With Schizophrenia, Depression, and Bipolar Disorder. JAMA Psychiatry. 2023;80(3):250-259. [38] MA C, ZHANG D, MA Q, et al. Arbutin inhibits inflammation and apoptosis by enhancing autophagy via SIRT1. Adv Clin Exp Med. 2021;30(5):535-544. [39] WANG X, RIBEIRO C, NILSSON A, et al. Oxytocin Receptor Expression and Activation in Parasympathetic Brainstem Cardiac Vagal Neurons. eNeuro. 2025;12(8):ENEURO.0204-25.2025. [40] CHEN B, CHEN Y, HE Z, et al. S146 and M148 within the mature chain domain of PSMB4 are crucial for degrading PRRSV nsp1α. Cell Mol Life Sci. 2025; 82(1):148. [41] LV J, LIU G, WANG Z, et al. Internalized polystyrene nanoplastics trigger testicular damage and promote ferroptosis via CISD1 downregulation in mouse spermatocyte. J Nanobiotechnology. 2025;23(1):537. [42] VERONA F, DI BELLA S, SCHIRANO R, et al. Cancer stem cells and tumor-associated macrophages as mates in tumor progression: mechanisms of crosstalk and advanced bioinformatic tools to dissect their phenotypes and interaction. Front Immunol. 2025;16:1529847. [43] SI S, LIU H, XU L, et al. Identification of novel therapeutic targets for chronic kidney disease and kidney function by integrating multi-omics proteome with transcriptome. Genome Med. 2024;16(1):84. [44] GERARDS WL, HOP FW, HENDRIKS IL, et al.Cloning and expression of a human pro(tea)some beta-subunit cDNA: a homologue of the yeast PRE4-subunit essential for peptidylglutamyl-peptide hydrolase activity. FEBS Lett. 1994;346(2-3):151-155. [45] YANG C, YU P, YANG F, et al. PSMB4 inhibits cardiomyocyte apoptosis via activating NF-κB signaling pathway during myocardial ischemia/reperfusion injury. J Mol Histol. 2021;52(4):693-703. [46] ZHANG X, LIN D, LIN Y, et al. Proteasome beta-4 subunit contributes to the development of melanoma and is regulated by miR-148b. Tumour Biol. 2017;39(6):1010428317705767. [47] WANG H, HE Z, XIA L, et al. PSMB4 overexpression enhances the cell growth and viability of breast cancer cells leading to a poor prognosis. Oncol Rep. 2018;40(4):2343-2352. [48] LI X, SHEN A, ZHAO Y, et al. Mendelian Randomization Using the Druggable Genome Reveals Genetically Supported Drug Targets for Psychiatric Disorders. Schizophr Bull. 2023;49(5):1305-1315. [49] LIU J, CHENG Y, LI M, et al. Genome-wide Mendelian randomization identifies actionable novel drug targets for psychiatric disorders. Neuropsychopharmacology. 2023;48(2):270-280. [50] HU J, FU J, CAI Y, et al. Bioinformatics and systems biology approach to identify the pathogenetic link of neurological pain and major depressive disorder. Exp Biol Med (Maywood). 2024;249:10129. [51] DATTILO V, AMATO R, PERROTTI N, et al. The Emerging Role of SGK1 (Serum- and Glucocorticoid-Regulated Kinase 1) in Major Depressive Disorder: Hypothesis and Mechanisms. Front Genet. 2020; 11:826. [52] JI D, LIU W, CUI X, et al. A receptor-like kinase SlFERL mediates immune responses of tomato to Botrytis cinerea by recognizing BcPG1 and fine-tuning MAPK signaling. New Phytol. 2023;240(3):1189-1201. [53] BALHARA P, SHARMA S, VASUDEVA N, et al. Modulation of Nrf-2/HO-1/HIF-1α/TFAM pathways by Arbutin in rat model of cerebral ischemic stroke. Mol Cell Neurosci. 2025;134:104034. [54] JAIN A, GOEL V, SONI S, et al. Comparative Efficacy and Toxicity of Modified FOLFIRINOX and Gemcitabine Plus Nab-Paclitaxel in Advanced Pancreatic Ductal Adenocarcinoma: A Real-World Retrospective Analysis. Cureus. 2025;17(7): e87389. [55] ZHOU J, HU Y, JING P. Antidepressant-Like Effect of Saffron (Crocus sativus L.) in Mice Exposed to Chronic Unpredictable Mild Stress via Attenuating Neuroinflammation and Recovering Neuroplasticity. Mol Nutr Food Res. 2025:e70201. doi: 10.1002/mnfr.70201. [56] SUDHOLZ H, MENG X, PARK HY, et al. Core fucosylation of IL-2RB is required for natural killer cell homeostasis. Cell Rep. 2025;44(8):116101. [57] VOLNIN A, PARSIKOV A, TSYBULKO N, et al. Ergot alkaloid control in biotechnological processes and pharmaceuticals (a mini review). Front Toxicol. 2024;6:1463758. |
| [1] | Guo Ying, Tian Feng, Wang Chunfang. Potential drug targets for the treatment of rheumatoid arthritis: large sample analysis from European databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1549-1557. |
| [2] | Wu Zhilin, , He Qin, Wang Pingxi, Shi Xian, Yuan Song, Zhang Jun, Wang Hao . DYRK2: a novel therapeutic target for rheumatoid arthritis combined with osteoporosis based on East Asian and European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1569-1579. |
| [3] | Liu Hongtao, Wu Xin, Jiang Xinyu, Sha Fei, An Qi, Li Gaobiao. Causal relationship between age-related macular degeneration and deep vein thrombosis: analysis based on genome-wide association study data [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1602-1608. |
| [4] | Gao Zengjie, , Pu Xiang, Li Lailai, Chai Yihui, Huang Hua, Qin Yu. Increased risk of osteoporotic pathological fractures associated with sterol esters: evidence from IEU-GWAS and FinnGen databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1302-1310. |
| [5] | Liu Fengzhi, Dong Yuna, Tian Wenyi, Wang Chunlei, Liang Xiaodong, Bao Lin. Gene-predicted associations between 731 immune cell phenotypes and rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1311-1319. |
| [6] | Zhang Cuicui, Chen Huanyu, Yu Qiao, Huang Yuxuan, Yao Gengzhen, Zou Xu. Relationship between plasma proteins and pulmonary arterial hypertension and potential therapeutic targets [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1331-1340. |
| [7] | Zeng Hao, Sun Pengcheng, Chai Yuan, Huang Yourong, Zhang Chi, Zhang Xiaoyun. Association between thyroid function and osteoporosis: genome-wide data analysis of European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1019-1027. |
| [8] | Rong Xiangbin, , Zheng Haibo, Mo Xueshen, Hou Kun, Zeng Ping, . Plasma metabolites, immune cells, and hip osteoarthritis: causal inference based on GWAS data from European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1028-1035. |
| [9] | He Qiwang, , , Chen Bo, Liang Fuchao, Kang Zewei, Zhou Yuan, Ji Anxu, Tang Xialin, . Relationship between Alzheimer’s disease and sarcopenia and body mass index: analysis of GWAS datasets for European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1036-1046. |
| [10] | Ding Yu, Chen Jingwen, Chen Xiuyan, Shi Huimin, Yang Yudie, Zhou Meiqi, Cui Shuai, . Circulating inflammatory proteins and myocardial hypertrophy: large sample analysis of European populations from GWAS Catalog and FinnGen databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1047-1057. |
| [11] | Zhao Feifan, Cao Yujing. An artificial neural network model of ankylosing spondylitis and psoriasis shared genes and machine learning-based mining and validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 770-784. |
| [12] | Liu Chu, Qiu Boyuan, Tong Siwen, He Linyuwei, Chen Haobo, Ou Zhixue. A genetic perspective reveals the relationship between blood metabolites and osteonecrosis: an analysis of information from the FinnGen database in Finland [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 785-794. |
| [13] | Huang Zhe, Shang Baoling, Yao Gengzhen, Pan Guangming. Association between immune cells and cardiovascular disease risk: a genome-wide association study in European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(23): 5992-5999. |
| [14] | Zhang Zheng, Zhang Yibo, Xu Bin, Yan Shichao, Guo Hui. Sarcopenia and non-alcoholic fatty liver disease: analysis of the gut microbiota [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(23): 6000-6009. |
| [15] | Yin Xingxiao, Jiang Yang, Song Yanping, Yao Na, Shen Zhen, Li Yanqi, Song Yueyu, Peng Hao, Chen Qigang. Association between sarcopenia and osteoporosis: a genome-wide data analysis in European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(23): 6030-6039. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||