Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (4): 1047-1057.doi: 10.12307/2025.944
Previous Articles Next Articles
Circulating inflammatory proteins and myocardial hypertrophy: large sample analysis of European populations from GWAS Catalog and FinnGen databases
Ding Yu1, Chen Jingwen2, Chen Xiuyan1, Shi Huimin1, Yang Yudie1, Zhou Meiqi3, Cui Shuai1, 3
- 1College of Acupuncture and Massage, Anhui University of Chinese Medicine, Hefei 230001, Anhui Province, China; 2First Affiliated Hospital of Nanjing Medical University, Nanjing 210000, Jiangsu Province, China; 3Anhui Provincial Key Laboratory of Meridian-Organ Correlation, Hefei 230001, Anhui Province, China
-
Received:2024-10-15Accepted:2024-12-12Online:2026-02-08Published:2025-05-23 -
Contact:Cui Shuai, MD, Profesosr, College of Acupuncture and Massage, Anhui University of Chinese Medicine, Hefei 230001, Anhui Province, China; Anhui Provincial Key Laboratory of Meridian-Organ Correlation, Hefei 230001, Anhui Province, China -
About author:Ding Yu, Master candidate, College of Acupuncture and Massage, Anhui University of Chinese Medicine, Hefei 230001, Anhui Province, China -
Supported by:National Key Research & Development Program, No. 2022YFC3500502 (to ZMQ); Key Project for Cultivating Outstanding Young Teachers, No. YQZD2023046 (to CS)
CLC Number:
Cite this article
Ding Yu, Chen Jingwen, Chen Xiuyan, Shi Huimin, Yang Yudie, Zhou Meiqi, Cui Shuai, . Circulating inflammatory proteins and myocardial hypertrophy: large sample analysis of European populations from GWAS Catalog and FinnGen databases[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1047-1057.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
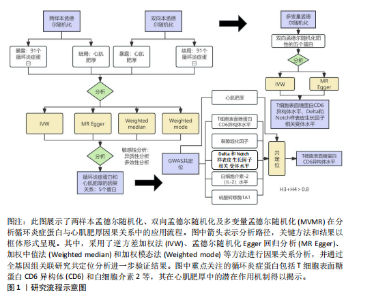
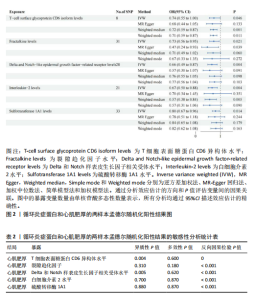
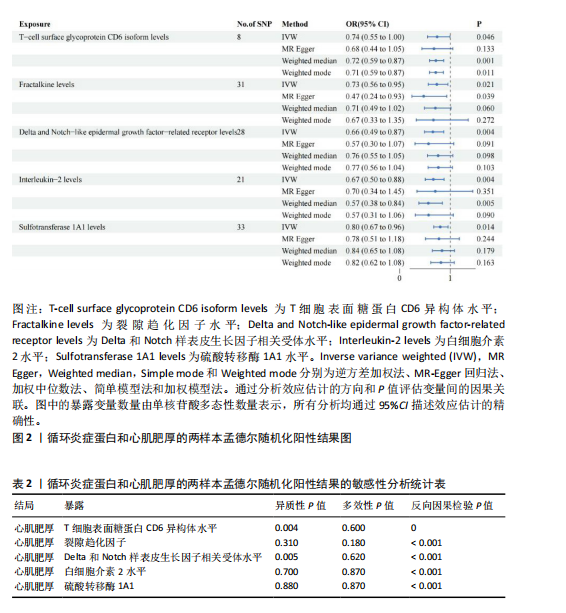
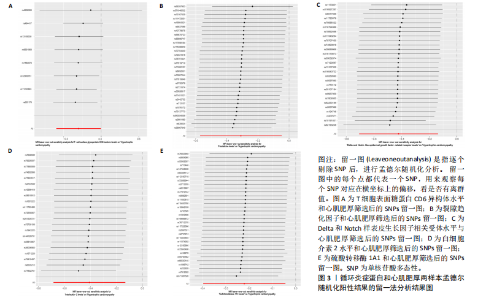
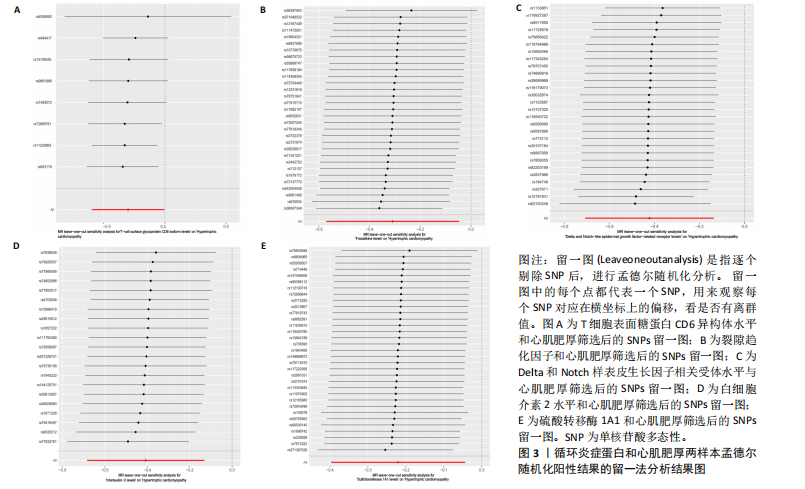
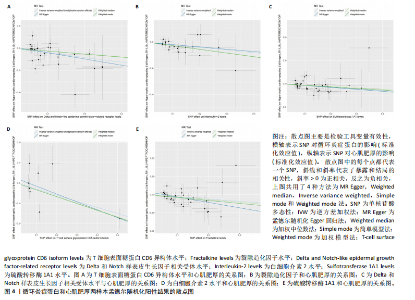
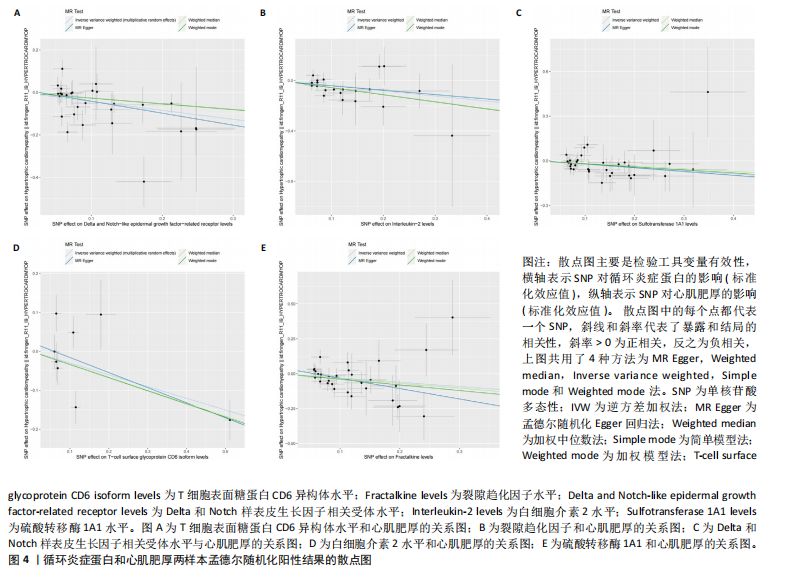
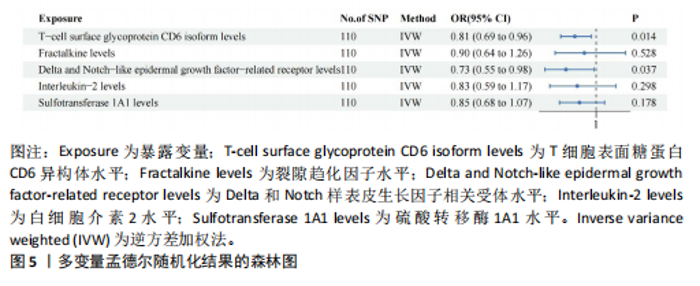
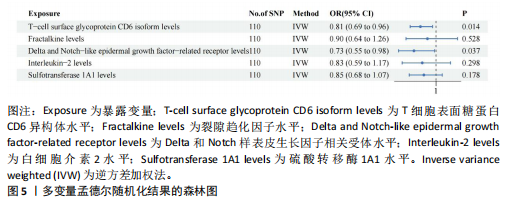
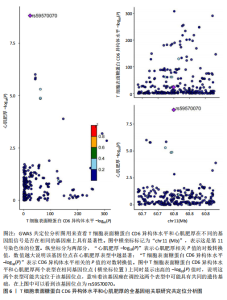
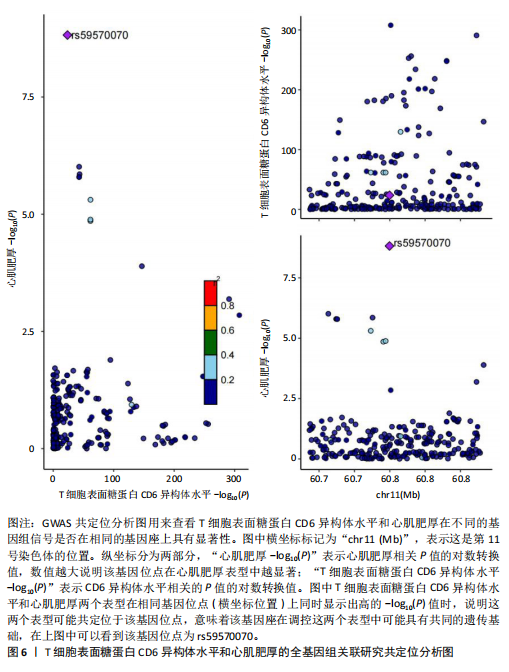
2.1 研究流程 此研究的暴露——循环炎症蛋白来自欧 洲 生 物 信 息 研 究 所 GWAS 目录数据库,研究人员采取用Olink Target平台对 14 824 名参与者中的 91 种血浆蛋白进行了全基因组蛋白质数量性状位点 (pQTL) 研究,并采用精准蛋白质组学技术对样本进行扫描,排除了脑源性神经营养性因子。同时用当下最新的第11版芬兰数据库中的心肌肥厚作为结局,共涵盖了453 733名参与者。该心肌肥厚数据库按照国际疾病分类(ICD-10)I42.1型,I42.2为条件,筛选出了具有显著指标的1 245例患者,并将他们的SNP作为结局变量。通过选择合适的条件筛选出SNP后,再将他们进行孟德尔随机化分析,最后通过留一法和MR-PRESSO方法进行筛选,此次未筛选出异常值。随后进行了敏感性和随机性分析,保证了所有结果的稳健性。最后还针对孟德尔随机化的结果进行了反向孟德尔随机化和多变量孟德尔随机化以及共定位分析,见图1。 2.2 工具变量的筛选结果 文章对此研究的工具变量进行严格的筛选,以P显著性为1×10-2为标准纳入与之相关的工具变量,其中在阳性的结果中,分别纳入了8个SNP作为T细胞表面糖蛋白CD6异构体水平的工具变量,31个SNP作为裂隙趋化因子的工具变量,28个SNP作为Delta和Notch样表皮生长因子相关受体水平的工具变量,21个SNP作为白细胞介素2水平的工具变量,33个SNP作为白细胞介素2的工具变量。查阅PhenoScanner网站后,发现并没有与心肌肥厚显著相关的SNP。同时,所有以上SNP的F值统计量均大于10 (F > 10),表明以上工具变量具有强烈相关性,文章中出现弱工具变量导致研究结果偏差的可能性极小。 2.3 孟德尔随机化分析结果 通过孟德尔随机化法对91种循环炎症蛋白和心肌肥厚疾病进行分析。有以下5种蛋白表明在遗传学上可能与心肌肥厚存在关系,其中 T细胞表面糖蛋白CD6异构体水平(IVW:P=0.046,OR=0.74,95%CI:0.55-1.00),裂隙趋化因子(又称为趋化因子裂隙趋化因子1)水平(IVW:P=2.1×10-2,OR=0.73,95%CI:0.556-0.95),Delta和Notch样表皮生长因子相关受体水平(IVW:P=3.7×10-4,OR=0.66,95%CI:0.49-0.87),白细胞介素2水平(IVW:P= 3.8×10-3,OR=0.667,95%CI:0.50-0.88), 硫酸转移酶1A1(IVW:P= 1.42×10-2,OR=0.80,95%CI:0.67-0.96)。文章还利用其他几种孟德尔随机化方法如MR Egger回归、加权中位数法和加权众数对结果进行了验证,他们的Beta值为同一指向,同一暴露的不同仿佛均显示该暴露与心肌肥厚呈正相关或负相关,这意味这结果稳健可靠,可作为遗传学上判断二者相关性的依据,见图2。在反向孟德尔随机化中,并未发现任何心肌肥厚作为暴露的情况下,为阳性结局的循环炎症蛋白。 2.4 敏感性分析结果 为加强结果的稳定性,使研究成果更有说服力,在此研究中还对结果进行了敏感性分析。MR-Egger截距检验表明文章不存在异质性和水平多效性(P > 0.05)。经Cochran’s Q检验,除Delta和Notch样表皮生长因子相关受体水平外其他结果P值均大于0.05,说明无异质性。而Delta和Notch样表皮生长因子相关受体存在异质性可能是因为个别样本量相互作用导致,但对结果并无影响。通过MR-PRESSO方法未检测到异常值(P > 0.05),更加肯定本结果在MR-PRESSO方法层面上无多效性和异质性,表明结果可靠,无其他混杂因素影响。Steiger 检验的结果P均小于0.05为TRUE,说明阳性暴露对结果均为单向阳性作用,这一点和双向孟德尔随机化结果相辅相成。留一分析和漏斗图未显示异常,说明不存在偏移的SNP,见图3,4。敏感性分析肯定了结果的稳定性和可靠性,见表2。 2.5 多变量孟德尔随机化分析结果 因为此研究采用的是批量两样本孟德尔随机化分析,为了排除其他阳性暴露的影响更准确地评估每个暴露因子对某一结局的独立因果效应,文章还做了多变量孟德尔随机化分析,见图5。基于两样本孟德尔随机化分析,对5个阳性暴露进行了分析,结果如下:T细胞表面糖蛋白CD6异构体水平(IVW:P=1.39×10-2,OR=0.81,95%CI:0.69-0.96),裂隙趋化因子水平(IVW:P=5.28×10-2,OR=0.90,95%CI:0.64-1.26),Delta和Notch样表皮生长因子相关受体水平(IVW:P=3.7×10-2, OR=0.73,95%CI:0.55-0.98),白细胞介素2水平(IVW:P=0.30,OR=0.83,95%CI:0.59-1.17),硫酸转移酶1A1 (IVW:P=0.18,OR=0.85,95%CI:0.68-1.07)。从以上结果可知,T细胞表面糖蛋白CD6异构体水平和Delta和Notch样表皮生长因子相关受体水平在多变量孟德尔随机化中为阳性结果,说明在其他的阳性暴露影响下,这二者对心肌肥厚依然有相当大的影响。而Delta和Notch样表皮生长因子相关受体水平、白细胞介素2水平和硫酸转移酶1A1在多变量结果中呈阴性,说明这三者对心肌肥厚具有影响,但在其他阳性暴露的共同影响下,三者的影响没那么显著。 2.6 共定位分析结果 为了找出在阳性结果中最显著的SNP基因和二者在基因范围上的关系,还进行了全基因组关联研究共定位分析,其中共定位的结果如下:T细胞表面糖蛋白CD6异构体水平H3+H4=0.96,其中H3=0.89, H4=0.07, 裂隙趋化因子水平H3+H4=0.11,Delta和Notch样表皮生长因子相关受体水平H3+H4=0.04,白细胞介素2水平H3+H4=0.01,硫酸转移酶1A1 H3+H4=0.03。从以上结果可知,几个阳性结果在共定位中有显著意义的为T细胞表面糖蛋白CD6异构体水平。 T细胞表面糖蛋白CD6异构体水平和心肌肥厚的共定位结果中,H3=0.89意味着T细胞表面糖蛋白CD6异构体水平与心肌肥厚在该区域内的SNP位点显著相关,但是是不同的因果变异位点。并且文章测得它的最显著的SNP为rs59570070,见图6,最显著的SNP是指与表型关联最强的SNP,通常是指可能和表型相关的关键位点。因此rs59570070可用于识别T细胞表面糖蛋白CD6异构体和心肌肥厚相关基因区域的潜在基因位点,帮助探寻二者在机制上的联系[44]。 2.7 假设验证结果 遗传变异通过影响循环炎症蛋白水平,与心肌肥厚的风险相关。文章的研究结果充分印证了这一假设,得出了循环炎症蛋白在遗传学上与心肌肥厚存在一定的关联,并且筛选出了5种具有显著意义的循环炎症蛋白,分别是T细胞表面糖蛋白CD6异构体水平、裂隙趋化因子、Delta和Notch样表皮生长因子相关受体水平、白细胞介素2水平和磺基转移酶。这也证实了先前的假设,即在基于遗传学工具变量,推断炎症蛋白的升高是会增加心肌肥厚的风险,验证了炎症蛋白是心肌肥厚的因果因素。这一假设的验证将为炎症蛋白在心肌疾病中的作用提供新的见解,并可能为预防和治疗心肌肥厚提供潜在的生物学靶点。药物或生活方式干预措施可以针对炎症反应进行管理,从而降低心肌肥厚的风险,该发现可能进一步拓展心血管疾病的治疗策略。"

| [1] D’AMATO G, LUXÁN G, DE LA POMPA JL. Notch signalling in ventricular chamber development and cardiomyopathy. FEBS J. 2016;283(23):4223-4237. [2] FONSECA FA, IZAR MC. Role of inflammation in cardiac remodeling after acute myocardial infarction. Front Physiol. 2022;13:927163. [3] DUBOIS-DERUY E, PEUGNET V, TURKIEH A, et al. Oxidative stress in cardiovascular diseases. Antioxidants (Basel). 2020;9(9): 864-864. [4] NAUSHAD S, GAUCHER J, MEZDARI Z, et al. Chronic intermittent hypoxia triggers cardiac fibrosis: role of epididymal white adipose tissue senescent remodeling? Acta Physiol(Oxf). 2024;240(11):e14231. [5] SHIMADA YJ, RAITA Y, LIANG LW, et al. Comprehensive proteomics profiling reveals circulating biomarkers of hypertrophic cardiomyopathy. Circ Heart Fail. 2021;14(7): e007849. [6] DINA RADENKOVIĆ, SAURABH CHAWLA, GIANNI BOTTA, et al. Advanced cardiometabolic & inflammatory markers for prediction of cardiovascular disease and cancer. Eur Heart J. 2020. doi:10.1093/ehjci/ehaa946.2921. [7] ARNOLD N, BLAUM C, GOßLING A, et al. C-reactive protein modifies lipoprotein(a)-related risk for coronary heart disease: the BiomarCaRE project. Eur Heart J. 2024; 45(12):1043-1054. [8] MORITA H, REHM HL, MENESSES A, et al. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358(18):1899-1908. [9] JI M, LIU Y, ZUO Z, et al. Downregulation of amphiregulin improves cardiac hypertrophy via attenuating oxidative stress and apoptosis. Biol Direct. 2022;17(1):21. [10] BAZGIR F, NAU J, NAKHAEI‐RAD S, et al. The microenvironment of the pathogenesis of cardiac hypertrophy. Cells. 2023;12(13):1780-1780. [11] MARON BJ, OMMEN SR, SEMSARIAN C, et al. Hypertrophic cardiomyopathy: present and future, with translation into contemporary cardiovascular medicine. J Am Coll Cardiol. 2014;64(1):83-99. [12] ELLIOTT PM, ANASTASAKIS A, BORGER MA, et al. 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the task force for the diagnosis and management of hypertrophic cardiomyopathy of the european society of cardiology (ESC). Eur Heart J. 2014;35(39):2733-2779. [13] CHENG Z, FANG T, HUANG J, et al. Hypertrophic cardiomyopathy: from phenotype and pathogenesis to treatment. Front Cardiovasc Med. 2021;8:722340. [14] LOPES LR, AUNG N, VAN DUIJVENBODEN S, et al. Prevalence of hypertrophic cardiomyopathy in the UK Biobank Population. JAMA Cardiol. 2021;6(7):852-854. [15] DAVEY SMITH G, HEMANI G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89-R98. [16] WANG Y, SHEN H. Challenges and factors that influencing causal inference and interpretation, based on Mendelian randomization studies. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(8):1231-1236. [17] GUO Z, WANG Z, GAO Z, et al. The status and trends of mitochondrial dynamics research: a global bibliometric and visualized analysis. J Bioenerg Biomembr. 2023;55(1):43-57. [18] ZHAO JH, STACEY D, ERIKSSON N, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol. 2023;24(9):1540-1551. [19] LOUISE ACM, GEORGE DAVEY S, KATE T. The global randomization test: a Mendelian randomization falsification test for the exclusion restriction assumption. medRxiv (Cold Spring Harbor Laboratory). 2022. doi:10.1101/2022.05.03.22274459. [20] BELL KJ, LOY C, CUST AE, et al. Mendelian randomization in cardiovascular research: establishing causality when there are unmeasured confounders. 2021;14(1): e005623. [21] QINGYUAN Z, JINGSHU W, GIBRAN H, et al. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann Stat. 2020. doi:10.1214/19-aos1866. [22] ZHIHAN X, ZICHEN W, TONGYU Z, et al. Bidirectional Mendelian randomization analysis of the genetic association between primary lung cancer and colorectal cancer. J Transl Med. 2023;21(1):722. [23] STALEY JR, BLACKSHAW J, KAMAT MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207-3209. [24] BURGESS S, BUTTERWORTH A, THOMPSON SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658-665. [25] FANYE W, MINGZHE C, MINGHUI Z, et al. Causal association between oral disease and chronic obstructive pulmonary disease: a Mendelian Randomization study. Res Square. 2023. doi:10.21203/rs.3.rs-3179826/v1. [26] BOWDEN J, DAVEY SMITH G, BURGESS S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. [27] BOWDEN J, DAVEY SMITH G, HAYCOCK PC, et al. Consistent estimation in Mendelian Randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016; 40(4):304-314. [28] HARTWIG FP, DAVEY SMITH G, BOWDEN J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985-1998. [29] ZHUANG X, YIN Q, YANG R, et al. Causal pathways in lymphoid leukemia: the gut microbiota, immune cells, and serum metabolites. Front Immunol. 2024;15: 1437869. [30] EVANS DM, DAVEY SMITH G. Mendelian Randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet. 2015; 16(1):327-350. [31] REBECCA CR, GEORGE DAVEY S. Commentary: Orienting causal relationships between two phenotypes using bidirectional Mendelian randomization. Int J Epidemiol. 2019;48(3):907-911 [32] COHEN JF, CHALUMEAU M, COHEN R, et al. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J Clin Epidemiol. 2015;68(3):299-306. [33] ZHAO L, HU M, LI L. Identifying the genetic associations between diabetes mellitus and the risk of vitiligo. Clin Cosmet Investig Dermatol. 2024;17:2261-2271. [34] BOWDEN J, HEMANI G, DAVEY SMITH G. Invited commentary: detecting individual and global horizontal pleiotropy in mendelian randomization-a job for the humble heterogeneity statistic? Am J Epidemiol. 2018;187(12):2681-2685. [35] HEMANI G, TILLING K, DAVEY SMITH G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081. [36] SLOB EAW, BURGESS S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44(4):313-329. [37] FATIMA B, ASHISH P, DIPENDER G, et al. Disentangling the effects of traits with shared clustered genetic predictors using multivariable Mendelian randomization. Genet Epidemiol. 2022;46(7):415-429. [38] ELEANOR S, TOM GR, TIM M, et al. Estimation of causal effects of a time-varying exposure at multiple time points through multivariable mendelian randomization. PLoS Genet. 2022;18(7):e1010290-e1010290. [39] MARIE CS, CHIARA A, KAIDO L, et al. Quantifying the role of transcript levels in mediating DNA methylation effects on complex traits and diseases. Nat Commun. 2022;13(1):7559. [40] STEPHEN B, SIMON G. THOMPSON. Multivariable Mendelian Randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251-260. [41] ZUBER V, GRINBERG NF, GILL D, et al. Combining evidence from Mendelian randomization and colocalization: review and comparison of approaches. Am J Hum Genet. 2022;109(5):767-782. [42] CLAUDIA G, DAMJAN V, SCHADT EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383. [43] WEI‐MING S, XIAOJING G, MENG D, et al. Systematic druggable genome-wide Mendelian randomisation identifies therapeutic targets for Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2023;94(11):954-961. [44] MARK IM, GONÇALO RA, LON RC, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356-369. [45] MARTA C, MARIO M, FERNANDO A, et al. Relevance of cd6-mediated interactions in the regulation of peripheral T-cell responses and tolerance. Front Immunol. 2017;8:594. [46] Breuning J, Brown MH. T cell costimulation by CD6 is dependent on bivalent binding of a gads/slp-76 complex. Mol Cell Biol. 2017;37(11):e00071. [47] BUGHANI U, SAHA A, KURIAKOSE A, et al. T cell activation and differentiation is modulated by a CD6 domain 1 antibody Itolizumab. PLoS One. 2017;12(7): e0180088. [48] DA GLÓRIA VG, MARTINS DE ARAÚJO M, MAFALDA SANTOS A, et al. T cell activation regulates CD6 alternative splicing by transcription dynamics and SRSF1. J Immunol. 2014;193(1):391-399. [49] HENRIQUES SN, OLIVEIRA L, SANTOS RF, et al. CD6-mediated inhibition of T cell activation via modulation of Ras. Cell Commun Signal. 2022;20(1):184. [50] SALA V, GALLO S, LEO C, et al. Signaling to cardiac hypertrophy: insights from human and mouse RASopathies. Mol Med. 2012;18:938-947. [51] FOLEY CN, STALEY JR, BREEN PG, et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021; 12(1):764. [52] CHEN X, YANG Y, SUN S, et al. CX3C chemokine: hallmarks of fibrosis and ageing. Pharmacol Res. 2024;208:107348. [53] WANG W, HU YF, PANG M, et al. BMP and Notch signaling pathways differentially regulate cardiomyocyte proliferation during ventricle regeneration. Int J Biol Sci. 2021;17(9):2157-2166. [54] ZHANG Y, DONG Y, XIONG Z, et al. Sirt6-mediated endothelial-to-mesenchymal transition contributes toward diabetic cardiomyopathy via the Notch1 signaling pathway. Diabetes Metab Syndr Obes. 2020;13:4801-4808. [55] POL JG, CAUDANA P, PAILLET J, et al. Effects of interleukin-2 in immunostimulation and immunosuppression. J Exp Med. 2019.doi:10.1084/jem.20191247. [56] XIAO J, YU K, LI M, et al. The IL-2/anti-IL-2 complex attenuates cardiac ischaemia-reperfusion injury through expansion of regulatory T cells. Cell Physiol Biochem. 2017;44(5):1810-1827. [57] ZENG Z, YU K, CHEN L, et al. Interleukin-2/anti-interleukin-2 immune complex attenuates cardiac remodeling after myocardial infarction through expansion of regulatory T cells. J Immunol Res. 2016; 2016:8493767. [58] SAK K, EVERAUS H. Sulfotransferase 1a1 as a biomarker for susceptibility to carcinogenesis: from molecular genetics to the role of dietary flavonoids. Curr Drug Metab. 2016;17(6):528-541. [59] WATANABE K, ARUMUGAM S, SREEDHAR R, et al. Small interfering RNA therapy against carbohydrate sulfotransferase 15 inhibits cardiac remodeling in rats with dilated cardiomyopathy. Cell Signal. 2015;27(7):1517-1524. [60] NEMET I, FUNABASHI M, LI XS, et al. Microbe-derived uremic solutes enhance thrombosis potential in the host. mBio. 2023;14(6):e0133123. [61] LIM YJ, SIDOR NA, TONIAL NC, et al. Uremic toxins in the progression of chronic kidney disease and cardiovascular disease: mechanisms and therapeutic targets. Toxins (Basel). 2021;13(2):142. [62] HIDEKI F, YURIKO Y, YUSUKE Y, et al. Anti‐oxidative effect of AST‐120 on kidney injury after myocardial infarction. Br J Pharmacol. 2016;173(8):1302-1313. [63] XIAOJIAO Z, XIAO-WEI H, YANJUN J, et al. Impact of inflammation and anti-inflammatory modalities on diabetic cardiomyopathy healing: from fundamental research to therapy. Int Immunopharmacol. 2023;123:110747. |
| [1] | Wen Xiaolong, Weng Xiquan, Feng Yao, Cao Wenyan, Liu Yuqian, Wang Haitao. Effects of inflammation on serum hepcidin and iron metabolism related parameters in patients with type 2 diabetes mellitus: a meta-analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1294-1301. |
| [2] | Gao Zengjie, , Pu Xiang, Li Lailai, Chai Yihui, Huang Hua, Qin Yu. Increased risk of osteoporotic pathological fractures associated with sterol esters: evidence from IEU-GWAS and FinnGen databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1302-1310. |
| [3] | Liu Fengzhi, Dong Yuna, Tian Wenyi, Wang Chunlei, Liang Xiaodong, Bao Lin. Gene-predicted associations between 731 immune cell phenotypes and rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1311-1319. |
| [4] | Zhang Cuicui, Chen Huanyu, Yu Qiao, Huang Yuxuan, Yao Gengzhen, Zou Xu. Relationship between plasma proteins and pulmonary arterial hypertension and potential therapeutic targets [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1331-1340. |
| [5] | Zeng Hao, Sun Pengcheng, Chai Yuan, Huang Yourong, Zhang Chi, Zhang Xiaoyun. Association between thyroid function and osteoporosis: genome-wide data analysis of European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1019-1027. |
| [6] | Rong Xiangbin, , Zheng Haibo, Mo Xueshen, Hou Kun, Zeng Ping, . Plasma metabolites, immune cells, and hip osteoarthritis: causal inference based on GWAS data from European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1028-1035. |
| [7] | He Qiwang, , , Chen Bo, Liang Fuchao, Kang Zewei, Zhou Yuan, Ji Anxu, Tang Xialin, . Relationship between Alzheimer’s disease and sarcopenia and body mass index: analysis of GWAS datasets for European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1036-1046. |
| [8] | Zhao Feifan, Cao Yujing. An artificial neural network model of ankylosing spondylitis and psoriasis shared genes and machine learning-based mining and validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 770-784. |
| [9] | Liu Chu, Qiu Boyuan, Tong Siwen, He Linyuwei, Chen Haobo, Ou Zhixue. A genetic perspective reveals the relationship between blood metabolites and osteonecrosis: an analysis of information from the FinnGen database in Finland [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 785-794. |
| [10] | Zhang Yibo, Lu Jianqi, Mao Meiling, Pang Yan, Dong Li, Yang Shangbing, Xiao Xiang. Exploring the causal relationship between rheumatoid arthritis and coronary atherosclerosis: a Mendel randomized study involving serum metabolites and inflammatory factors [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-9. |
| [11] | Chen Jiayong, Tang Meiling, Lu Jianqi, Pang Yan, Yang Shangbing, Mao Meiling, Luo Wenkuan, Lu Wei, Zhou Jiatan. Based on Mendelian randomization, the causal relationship between 1400 metabolites and sarcopenia and the correlation analysis of cardiovascular disease were investigated [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-11. |
| [12] | Dong Tingting, Chen Tianxin, Li Yan, Zhang Sheng, Zhang Lei. Causal relationship between modifiable factors and joint sports injuries [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1953-1962. |
| [13] | Chen Shuai, Jin Jie, Han Huawei, Tian Ningsheng, Li Zhiwei . Causal relationship between circulating inflammatory cytokines and bone mineral density based on two-sample Mendelian randomization [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1556-1564. |
| [14] |
Zhao Wensheng, Li Xiaolin, Peng Changhua, Deng Jia, Sheng Hao, Chen Hongwei, Zhang Chaoju, He Chuan.
Gut microbiota and osteoporotic fractures #br#
#br#
[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1296-1304.
|
| [15] | Ma Haoyu, Qiao Hongchao, Hao Qianqian, Shi Dongbo. Causal effects of different exercise intensities on the risk of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1305-1311. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||